Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike (S) protein binds the host receptor angiotensin-converting enzyme 2 (ACE2). This binding promotes virus membrane fusion and causes host cell infection. The S protein is extensively glycosylated, i.e., covered by glycans or sugar chain structures. The effects of this glycosylation and their role in the pathological behavior of the S protein are unknown.
A team of investigators, led by Dr. Benjamin F. Mann, from Merck, USA have published a new study on the preprint server bioRxiv*. They have used a biochemical approach to control the glycosylation profile of the S protein receptor-binding domain (RBD) and to analyze the effect of different glycosylation patterns on receptor binding.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
S protein RBD glycosylation
The S protein is heavily glycosylated and these glycans shield around 40% of the protein surface. Glycosylation is important for viral pathobiology. It is required for virus integrity and infectivity. Glycans can effectively mask viral immunogenic epitopes from the host immune system.
The SARS-CoV-2 virus can camouflage its epitopes with host-derived glycans. While the S protein is heavily glycosylated, the RBD has only two glycosylation sites making it vulnerable to immune recognition. Therefore, the investigators speculated that, apart from glycan shielding, these RBD glycans may be involved in the S protein-ACE2 interactions.
Due to technical challenges in controlling the glycan structures, glycan biochemistry is predominantly studied using an in silico approach and experimental data is lacking. Therefore, this study employs a glycoengineering approach to harmonize the glycans of S protein RBD into controlled terminal glycan species. Furthermore, it also assesses the binding affinity between the glycoengineered RBD and human ACE2.
RBD glycan remodeling and ACE2 interaction
The researchers analyzed the S protein RBD glycans by glycan chromatography and mass spectrometry; remodeled the RBD enzymatically using glycoengineering enzymes; and measured ACE2 binding affinity by binding affinity analysis.
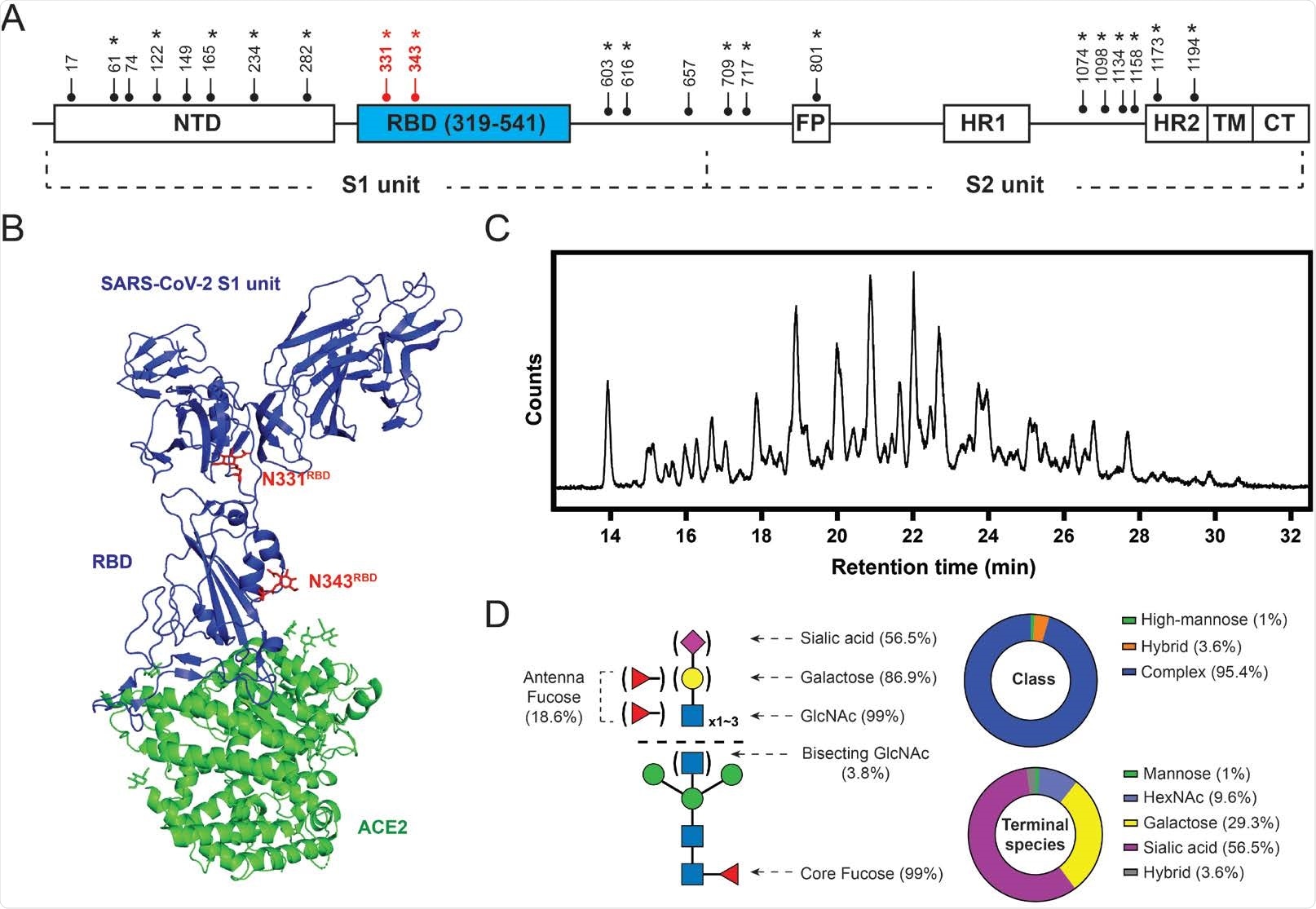
SARS-CoV-2 S protein glycosylation. (A) Glycosylation sites are distributed throughout the S protein structures. Star symbols (*) indicate conserved glycosylation sites with SARS-CoV S protein. NTD: N-terminal domain; RBD: receptor-binding domain; FP: fusion peptide; HR: heptad repeat; TM: transmembrane region; CT: cytoplasmic domain fusion. (B) Structure of the S protein S1-ACE2 complex with highlighted RBD glycosylation sites in red (Protein Data Bank (PDB): 7a92). (C) HILIC chromatogram of glycans collected from HEK293-expressed S protein RBD used in this study. (D) Composition of S protein RBD glycosylation described in relative abundance of glycoform(s) with each property. Left: graphic representative of the RBD glycopattern. Upper right: composition of glycan classes. Bottom right: composition of terminal saccharide species. 56.5% of the RBD glycan population contains at least one terminal sialic acid; 29.3% of the population exhibits at least one terminal galactose without having any sialic acid; 9.6% of the population terminating with GlcNAc without having any galactose and sialic acid.
RBD glycan regulatory role
Different glycoforms can have different regulatory effects on protein-protein interactions. Firstly, the researchers dissected the glycoforms of the recombinant RBD used in the study.
Next, they studied the interaction between glycoengineered RBD and ACE2. Glycans terminating with HexNAc, mannose and galactose have improved ACE2 binding affinity as compared to native RBD. Thus, these glycoforms can facilitate RBD-ACE2 interactions.
Conversely, glycans terminating with sialic acid have a weaker binding affinity to ACE2. Both, ACE2 surface and sialylated glycoforms are negatively charged and electrostatic repulsion may destabilize RBD-ACE2 interactions.
Furthermore, fucose in the RBD glycans does not have any effect on ACE2 binding affinity. This may be because fucose is close to the RBD peptide backbone and does not interact with ACE2.
ACE2 is also a glycosylated protein and it is known that ACE2 glycans interact with RBD glycans and the RBD protein backbone. Removal of RBD glycans reduces the RBD-ACE2 binding affinity. Removal of ACE2 glycans dramatically decreases the RBD-ACE2 binding affinity. Thus, the RBD glycans may interact with ACE2 glycans or ACE2 backbone to stabilize ACE2 binding.
The investigators also measured the binding affinity of an antibody named S309 and the different glycoengineered RBD. S309 is isolated from a SARS-CoV-infected patient, and it has a robust cross-neutralization activity towards SARS-CoV-2. S309 binds to RBD at an interface different from that of ACE2. The different glycoengineered RBD bind to S309 with similar affinities suggesting that RBD-S309 interaction may be dependent on the amino acid epitope of the RBD.
The investigators further probed the ACE2 binding with RBD bound to S309. ACE2 binds with similar affinities to different glycoengineered RBD bound to S309. Thus, S309 neutralizes the regulatory effects of the RBD glycans on RBD-ACE2 binding.
Advancing glycobiology and therapeutic strategies
Due to the rapid evolution of SARS-CoV-2 and the demand for effective therapies, it is essential to understand viral pathobiology.
Glycans confer structural and functional properties to a protein. The SARS-CoV-2 S protein is heavily glycosylated, barring the RBD, which has only two glycosylation sites. These RBD glycans are close to the binding interface of ACE2 and studying them has provided insights into the infection mechanism.
The neutrally charged RBD glycans can stabilize ACE2 binding, whereas the negatively charged sialic acids destabilize ACE2 binding.
The investigators suggest that strategies for regulation of RBD sialylation using drugs are worth investigating for SARS-CoV-2 treatment.
S309 neutralizes the regulatory effects of RBD glycans and prevents ACE2 binding. Since RBD glycosylation sites are conserved in all variants, the investigators propose that glycans can help to establish new strategies to deal with the emerging viral variant threat.
An important implication of the study is that it demonstrates the possibility to study different protein glycoforms biochemically. This glycoengineering approach can advance our knowledge in the field of glycobiology.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Hsu Y-P, Mukherjee D, Shchurik V, Makarov A, Mann BF. Structural remodeling of SARS-CoV-2 spike protein glycans reveals the regulatory roles in receptor binding affinity, bioRxiv, 2021:2021.08.26.457782. doi:10.1101/2021.08.26.457782, https://www.biorxiv.org/content/10.1101/2021.08.26.457782v1
- Peer reviewed and published scientific report.
Hsu, Yen-Pang, Martin Frank, Debopreeti Mukherjee, Vladimir Shchurik, Alexey Makarov, and Benjamin F Mann. 2022. “Structural Remodeling of SARS-CoV-2 Spike Protein Glycans Reveals the Regulatory Roles in Receptor-Binding Affinity.” Glycobiology 33 (2): 126–37. https://doi.org/10.1093/glycob/cwac077. https://academic.oup.com/glycob/article/33/2/126/6825274.