The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the pathogen behind the coronavirus disease (COVID-19) pandemic, continues to wreak havoc across the globe. To date, worldwide, infections have reached over 226 million, with over 4.65 million people having lost their lives.
For scientists to deal with the outbreak, they need to learn more about SARS-CoV-2's structure and how it infects the body.
Now, researchers at the Garvan Institute of Medical Research, Darlinghurst in Australia, CSIRO Data61, and the School of Biotechnology and Biomolecular Sciences (UNSW) demonstrated the most comprehensive analysis of the three-dimensional structure of SARS-CoV-2, providing a new insight into how the virus infects cells and replicates.
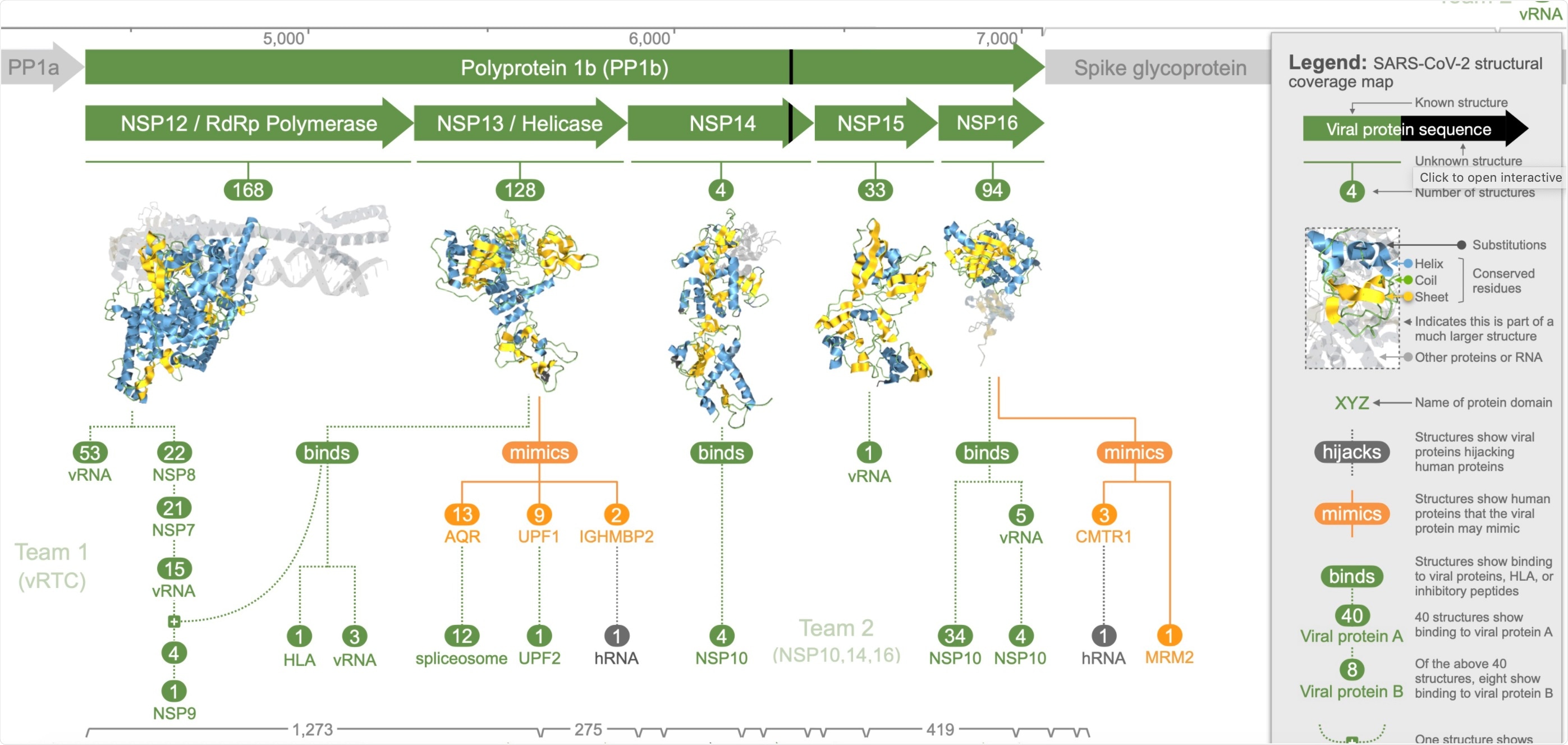
This Aquaria page summarises and maps all available 3D structural information for all proteins comprising the COVID-19 virus (SARS-CoV-2).
3D structures of all SARS-CoV-2 proteins
In the study, published in the Molecular Systems Biology journal, the team modeled 3D structures of all SARS-CoV-2 proteins, producing over 2,000 models that span 69 percent of the viral proteome. These models provide details not found anywhere else.
The different structures involved the coronavirus's 27 proteins. In addition, the analysis identified viral proteins that copy and hijack human proteins, allowing the virus to evade cell defenses and replicate to trigger an infection.
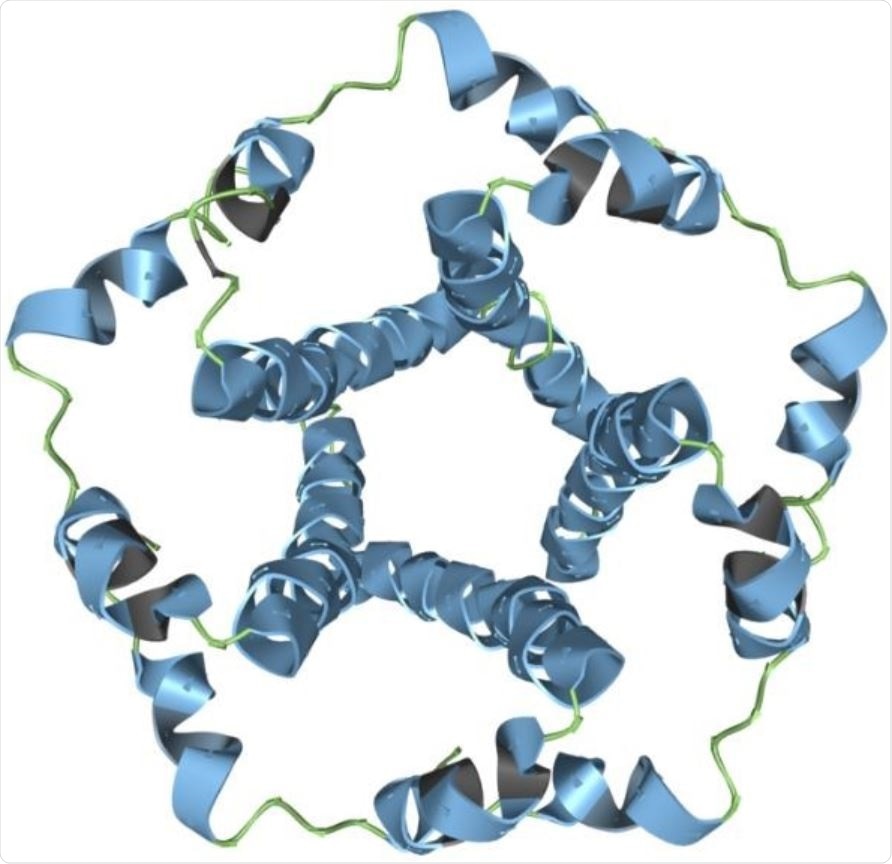
The SARS-CoV-2 envelope modeled in Aquaria. Image Credit: Garvan Institute of Medical Research
The study findings showed that about 6 percent of the proteome imitated human proteins, while approximately 7 percent was implicated in taking over human proteins, hence, reversing post-translational modifications, turning off host defenses, and blocking host translation.
The team made the structural models freely available at the Aquaria-COVID resource, a website designed to help researchers discover new potential targets on the virus to develop treatments or vaccines. The models can also help scientists study new emerging variants.
Switching focus on SARS-CoV-2
Amid the pandemic, many scientists have shifted their focus to SARS-CoV-2, including its structures like viral proteins. Specifically, they focused on the spike glycoprotein, the one responsible for binding to the human angiotensin-converting enzyme 2 (hACE2) receptor.
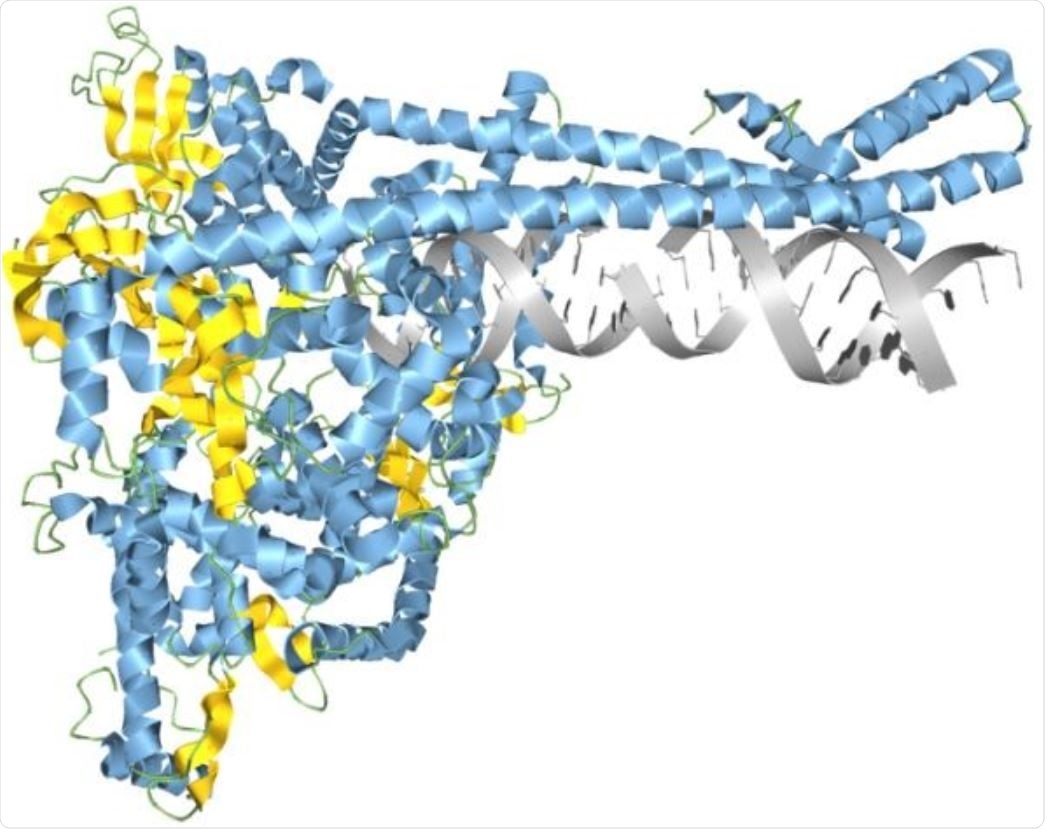
SARS-CoV-2 RNA synthesis complex modeled in Aquaria. Image Credit: Garvan Institute of Medical Research
Some modeling studies predicted 3D structures for the entire SARS-CoV-2 proteome, using technologies like AlphaFold, I-TASSER, Rosetta, SWISS-MODEL, and MODELLER. However, some of these models provided varying predictions, raising concerns about accuracy.
The scientists aimed to address such limitations through the use of a depth-based strategy that models all states with related 3D structures. The 3D structures of proteins provide atomic-resolution data on SARS-CoV-2's composition. This is essential to develop treatments and vaccines that target specific structures of the virus.
The study's findings showed three coronavirus proteins, NSP3, NSP16, and NSP13, mimicked human proteins. These proteins, called mimics, allow the virus to escape the immune system, affecting COVID-19 outcomes.
Further, the team also identified five other proteins, namely NSP1, NSP3, spike glycoprotein, ORF9b protein, and the envelope protein, collectively called hijackers, which invade and disrupt cell processes. As a result, the virus takes over to complete its life cycle and invade other cells.
The team also determined eight coronavirus proteins that can self-assemble called teams, providing new insights into how the virus replicates its genome.
"We found structural evidence of interaction between viral proteins for 29% of the viral proteome, comprising eight SARS-CoV-2 proteins," the team explained.
This leaves 14 of the 27 viral proteins in a category called suspects. These proteins are believed to play key roles in infection but have no structural evidence of interaction with other proteins.
In a nutshell, the study assembled information about the viral proteome's structure that was not available from other resources.
"Our analysis of these data has provided insight into how viral proteins self-assemble, how NSP3 and NSP13 may mimic human proteins, and how viral hijacking reverses post-translational modifications, blocks host translation, and disables host defenses," the researchers concluded in the study.
The study can help scientists develop effective treatment options and vaccines to fight off the pandemic.
Source:
Journal reference: