Since the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) originally emerged in Wuhan, China at the end of 2019, it has rapidly spread across populations and international boundaries. The resulting pandemic of the coronavirus disease 2019 (COVID-19) has caused severe repercussions on human health and well-being, including severe financial strain, unemployment, loss of jobs, and poor educational quality.
While the rollout of several vaccines promised to control the spread of the virus initially, the rapid emergence of new SARS-CoV-2 variants, some with markedly higher transmissibility and/or immune escape, has threatened the efficacy of current vaccines. A new study discusses the isolation of two variants of concern (VOCs) from an immunocompromised COVID-19 patient.
 Study: Divergence Of Delta and Beta Variants And SARS-Cov-2 Evolved in Prolonged Infection Into Distinct Serological Phenotypes. Image Credit: Cristi Savin / Shutterstock.com
Study: Divergence Of Delta and Beta Variants And SARS-Cov-2 Evolved in Prolonged Infection Into Distinct Serological Phenotypes. Image Credit: Cristi Savin / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
Current circulating VOCs include the Alpha, Beta, Gamma, and Delta variants of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the pathogen responsible for COVID-19. These variants not only inhibit neutralization by convalescent plasma containing antibodies induced by infection with the earlier strains of the virus, or antisera containing vaccine-induced antibodies but may also prevent the induction of an effective immune response by specific mutations. Moreover, these variants are often far more transmissible than the parental strain of SARS-CoV-2.
In terms of their ability to escape from immune neutralization, the most notable VOCs include the Beta, Gamma, and Delta variants. Each of these variants has substitutions in the spike protein at the epitopes, or antigenic sites, that make up the receptor-binding domain (RBD). The primary role of the RBD is to engage with the angiotensin-converting enzyme 2 (ACE2) on the host cell membrane to mediate viral entry.
The key mutations involved in neutralization escape include K417N/T, E484K, and N501Y mutations for the Beta and Gamma variants, with the Alpha also having the N501Y. Conversely, the Delta variant has the L452R and T478K substitutions in the RBD.
South Africa has witnessed three waves of COVID-19 so far, by the ancestral or wildtype virus (D614G), as well as the Beta and Delta variants, respectively.
Human immunodeficiency virus (HIV) infection can affect the immune response to other pathogens, likely in part through dysregulated CD4 T-helper cell function and a reduction in the number of these cells. While antiretroviral therapy (ART) for HIV avoids such severe depletion, non-compliance or drug resistance can allow the virus to continue to replicate, causing full-blown HIV over years.
In this case, SARS-CoV-2 can persist for months and evolve into distinct variants within the same immunocompromised host. The current study focused on such a scenario.
The patient had HIV but recovered clinically within 10 days. HIV was detectable in the blood for up to 190 days due to poor ART compliance. The patient subsequently developed SARS-CoV-2 infection, which was diagnosed 16 days after the onset of symptoms.
Study findings
Study enrollment occurred six days from diagnosis, with the CD4 T-cell count of less than 10. SARS-CoV-2 continued to be detected by polymerase chain reaction (PCR) up to day 216 from diagnosis and was isolated until day 190 from SARS-CoV-2 diagnosis.
Immunoglobulin G (IgG) antibodies to the RBD were borderline until day 190, but then increased and peaked at day 216.
Viral sequencing showed that various mutations occurred at various stages. The E484K mutation was first observed on day 6 and was subsequently replaced from day 71 with the F490S substitution. At this point, the K417T mutation was also detected. Much later, on day 190, the N501Y mutation arose.
Neutralization of the early samples by day 216 plasma showed high susceptibility for day 6 viral isolates, followed by day 20 and day 190 isolates. The same pattern was observed with convalescent plasma (CP) from other patients infected at the same time.
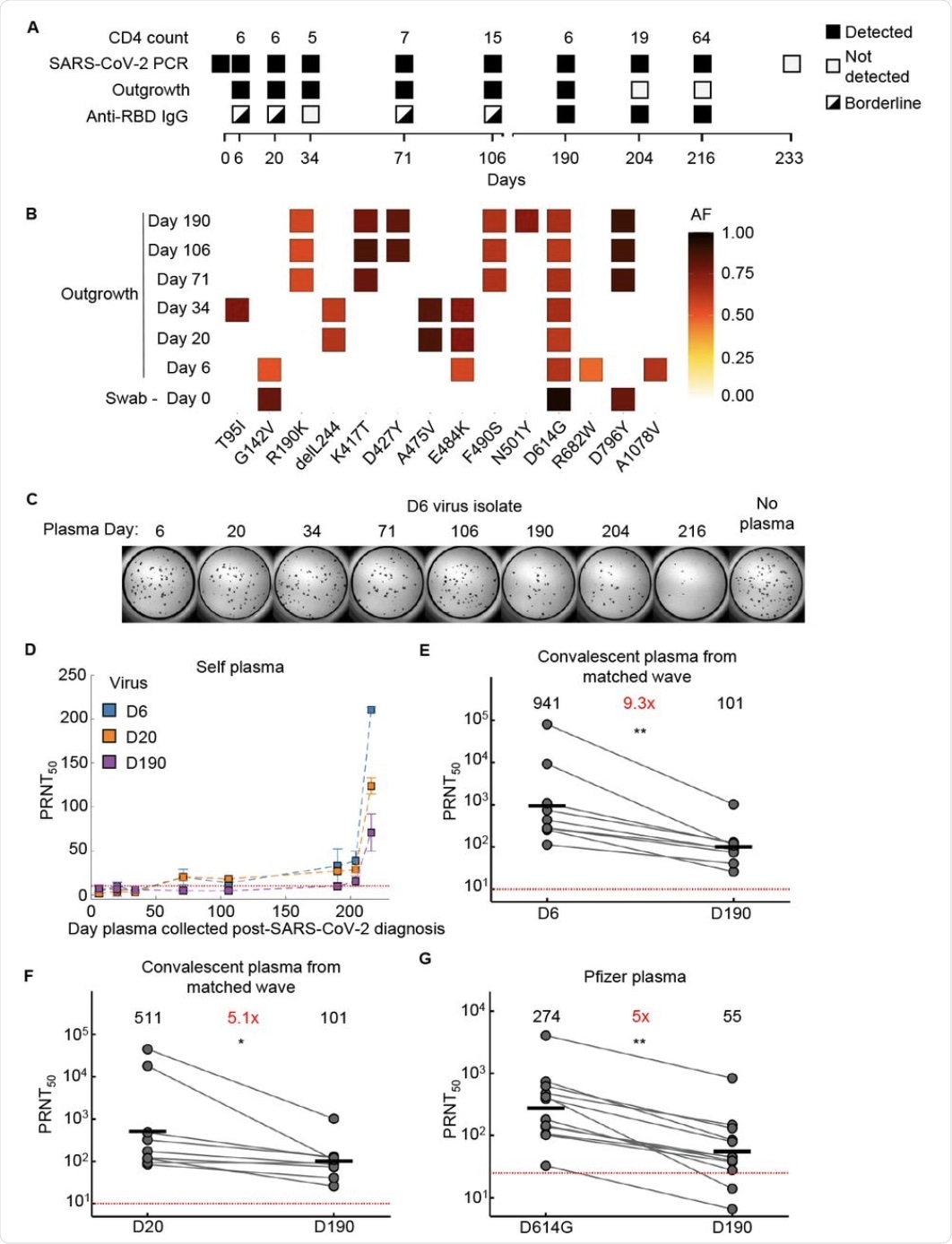 Evolution of SARS-CoV-2 with neutralization escape in a participant with advanced HIV
Evolution of SARS-CoV-2 with neutralization escape in a participant with advanced HIV
(A) Participant characteristics over 233 days from SARS-CoV-2 diagnosis including CD4 T cell count (cells/µL), presence or absence of SARS-CoV-2 by qPCR, success or failure of live virus outgrowth from swab sample, and presence or absence of anti-RBD IgG by ELISA. Because IgG levels were close to the background cutoff for some timepoints, they were marked as borderline. (B) Majority and minority SARS-CoV-2 genotypes in the swab (day 0) and outgrowth (day 6 to 190) sequences. X-axis lists substitutions and deletions in spike sequence. AF: allele frequency. (C) Live virus neutralization assay of the early day 6 (D6) SARS-CoV-2 virus isolate by self-plasma collected at days 6 to 216. Plasma dilution shown is 1:20. (D) Neutralization of D6, day 20 isolated (D20), and day 190 isolated (D190) virus by self-plasma collected days 6 to 216. (E) Neutralization of D6 virus relative to D190 virus by convalescent plasma of participants (n=8) infected by ancestral strains. Number in black above virus name is geometric mean of the PRNT50 and number in red represents fold-change of geometric mean between virus on the left and on the right of graph. (F) Neutralization of D20 isolate compared to D190 isolate by convalescent plasma from the same participants as in (E). (G) Neutralization of D190 isolate compared to ancestral virus (D614G) by Pfizer BNT162b2 elicited plasma (n=12). p-values are * <0.05; ** <0.01-0.001 as determined by the Wilcoxon rank sum test. Red horizontal line in panels D to G denotes most concentrated plasma tested.
The neutralization potency of CP against day 6 isolate was 9-fold higher compared to that of day 190, and five-fold higher than for day 20 isolate. With Pfizer vaccine-elicited antiserum, neutralization was five-fold less for the day 190 sample as compared to the earlier D614G strain.
“These results support evolution of escape from neutralization of D190 virus.”
Neutralization by D614G-elicited plasma was seven-fold less with the Beta VOC but not significantly less with the Delta strain. With the day 190 isolate, it was nine-fold less and thus more closely resembled the Beta variant.
With Beta-induced antibody-containing CP, Delta neutralization dropped by 12-fold more than the Beta variant. However, D190 was neutralized only 2-6-fold less powerfully.
The use of Delta-elicited antibody-rich CP showed steep declines in Beta or day 190 isolate neutralizing capacity at 34-fold and 27-fold less, respectively, as compared to that of the Delta variant. This surprising escape of the Beta variant prompted a pseudovirus neutralization assay for confirmation, with a similar 26-fold reduction in neutralization.
Implications
These results suggest that the Beta and Delta VOCs are separated by many antigenic epitope variations, from each other, but less so from the common ancestral strain. The antigenic distance from the parent is greater for the Beta VOC, thus explaining its greater immune escape capability due to the selection of mutations like E484K and K417N in response to the enrichment of antibodies targeting these sites in its environment.
The day 190 isolate showed antibody neutralization patterns similar to the Beta, but quite different from the Delta variant. This showed the most immune escape capability of all the variants isolated from this patient. Though two mutations in the RBD in this isolate were shared to the Gamma and Lambda strains, respectively, it behaves like the Beta variant in escaping neutralization from the Delta-elicited plasma.
This divergent strain may be the persistent exposure to low levels of antibodies against the RBD of the virus.
Earlier research by these authors has revealed the doubling of HIV-infected individuals with SARS-CoV-2 co-infection who are not ART-compliant between the first two waves of the pandemic in South Africa. Correspondingly, the number of people with HIV viremia also doubled, which was likely similar to the proportion of people with immunosuppression.
This dangerous situation is likely to yield many newer strains of SARS-CoV-2 if not corrected by encouraging increased adherence to ART.
Finally, the broad separation between the Beta and Delta variants, with the ancestral variant being intermediate between them, may herald selective immunity to the infecting variant while the individual remains vulnerable to other strains.
“This may imply that a vaccine which is broadly effective across variants should either be designed around the ancestral strain, or with all the diverging variants. The use of only one variant may generate immunity which is vulnerable to infection by another variant.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Cele, S., Karim, F., Lustig, G., et al. (2021). Divergence Of Delta and Beta Variants And SARS-Cov-2 Evolved in Prolonged Infection Into Distinct Serological Phenotypes. medRxiv. doi:10.1101/2021.09.14.21263564. https://www.medrxiv.org/content/10.1101/2021.09.14.21263564v1.
- Peer reviewed and published scientific report.
Cele, Sandile, Farina Karim, Gila Lustig, James Emmanuel San, Tandile Hermanus, Houriiyah Tegally, Jumari Snyman, et al. 2022. “SARS-CoV-2 Prolonged Infection during Advanced HIV Disease Evolves Extensive Immune Escape.” Cell Host & Microbe 30 (2): 154-162.e5. https://doi.org/10.1016/j.chom.2022.01.005. https://www.cell.com/cell-host-microbe/fulltext/S1931-3128(22)00041-5.