Accurate and real-time disease prediction is vital for the prevention and control of healthcare-related infections. Although contacts between individuals are primarily responsible for infection chains, most prediction frameworks do not capture the contact dynamics.
Researchers from the UK recently developed a real-time machine learning framework that uses dynamic patient contact networks to predict hospital-onset COVID-19 infections (HOCIs) at the patient level. They then tested and validated the framework on international multi-site datasets across various epidemic and endemic periods. This study can be found on the medRxiv* preprint server.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The framework extracts dynamic contact networks from hospital data collected routinely and combines them with clinical attributes and background contextual hospital data of the patient to predict the infection status of patients. The research team trained and tested the HOCI framework using over 50,000 patients admitted to a UK National Health Service (NHS) Trust hospital between 01 April 2020 and 01 April 2021, thus spanning COVID-19 surges 1 and 2 in the UK.
They then validated the HOCI framework by applying it to data from 40,057 inpatients gathered from a hospital site in Geneva during an epidemic surge and data from 43,375 inpatients collected from the same NHS Trust from a subsequent period after surge 2, when COVID-19 had become endemic.
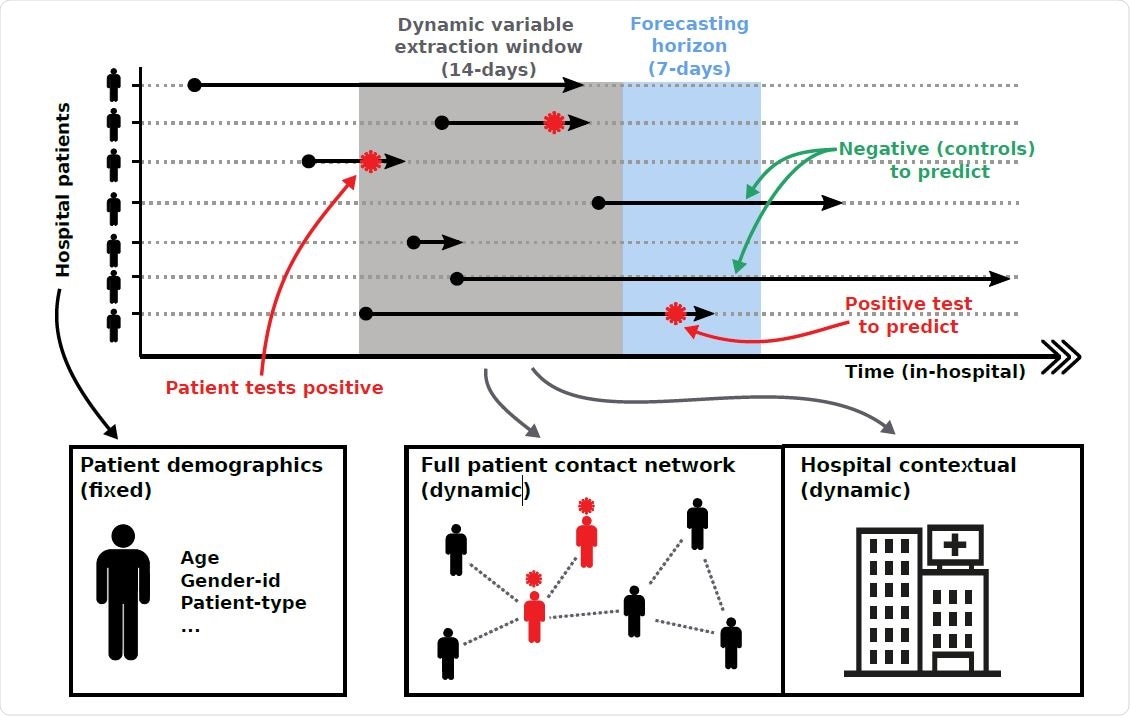
Overview of forecasting framework. Patient pathways are extracted from electronic health records which specify the locations each patient has visited over the duration of their hospital stay. Pathways are overlaid with COVID-19 testing results, capturing the space-time positions of patients that tested positive for COVID-19. Forecasting is based on extracting individual patient clinical variables (fixed) and hospital contextual variables (dynamic) during a defined time window, as well as variables capturing the centrality of a patient within the different contact networks (dynamic). We iterate variable extraction over multiple time windows and use the cumulative information for model training and predictions.
The HOCI framework demonstrated high predictive performance while using only contact network variables
Based on the training data from London spanning COVID-19 surges 1 and 2, the HOCI framework showed high predictive performance using all variables but was almost equally predictive using only contact network variables and more predictable than using only hospital contextual or patient clinical variables.
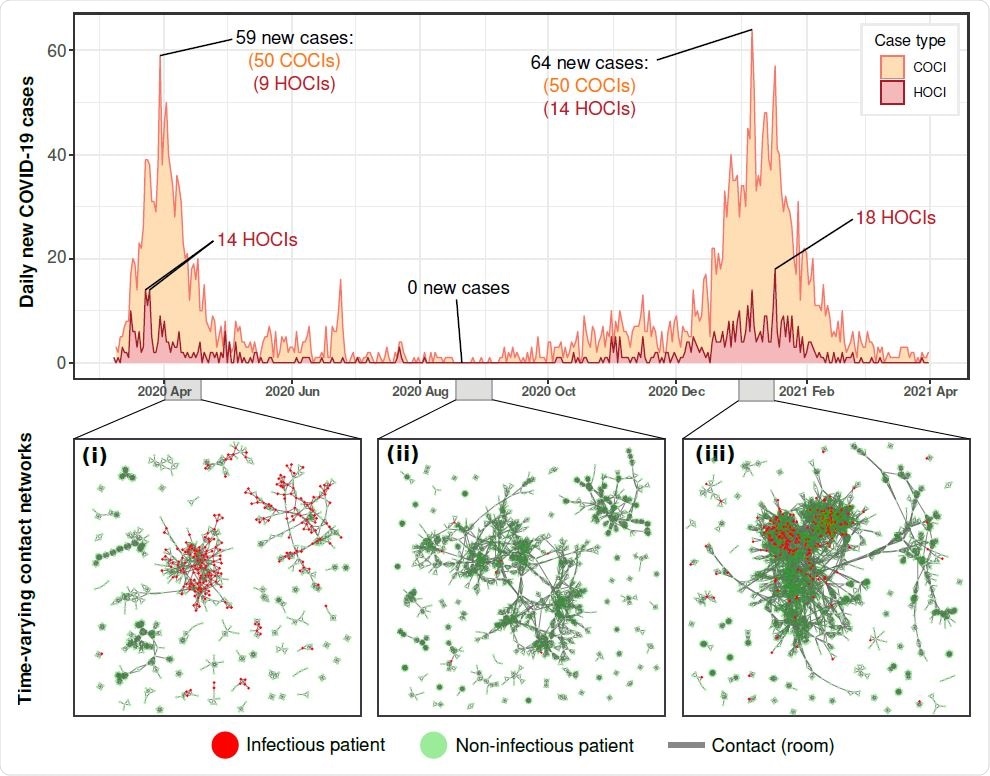
Background hospital infections and contact structure across the study period. Daily new COVID-19 positive patients within the hospital (COCI and HOCI) varied dramatically across the study period. Peaks of 59 and 64 cases were reached on March 30, 2020, and January 06, 2021 respectively, whilst dipping to zero new daily cases over days during July, August, September, and October. The patient contact network also varied across the study period, panels i-iii, with differences in connectivity and size of patient contact clusters between each of the infection surges and during the summer period.
The top 3 risk factors identified by the team comprised one hospital contextual variable, namely, background hospital COVID-19 prevalence, and two contact network variables, namely, the number of direct contacts of infectious patients and network closeness. Moreover, the addition of contact network variables boosted performance related to hospital contextual variables on both the UK and non-UK validation datasets.
“Our study highlights the predictive power that can be mined from networks of patient contacts to aid with personalized predictions of infectious disease in healthcare settings.”
Findings show that integration of patient contact networks in clinical care could improve infection prevention and help early diagnosis
According to the authors, this is the first study to predict individual patient HOCIs using routine patient and hospital data and dynamic contact networks. The research team combined machine learning with network analysis and developed a HOCI prediction framework with the help of routine electronic health data.
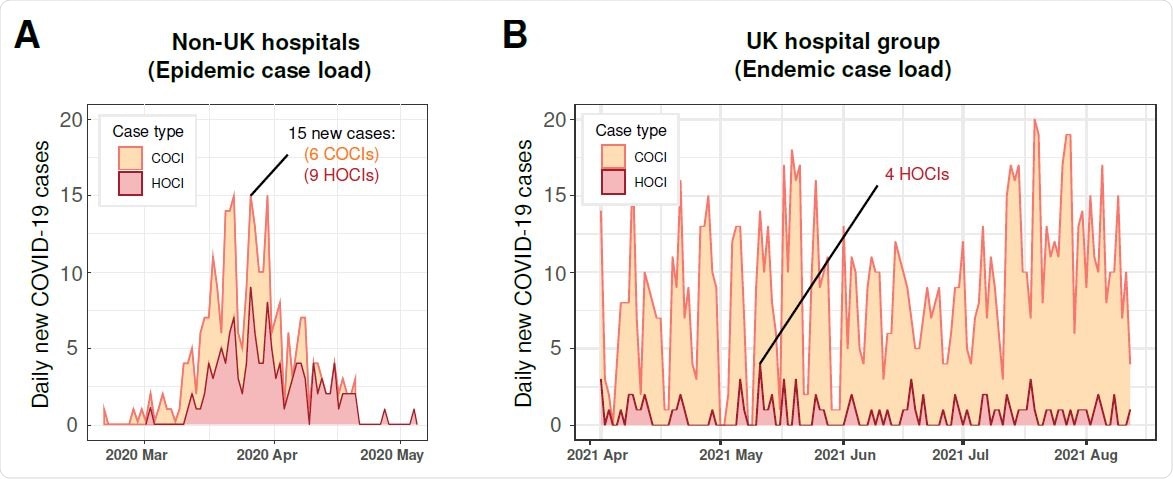
Epidemiology curves of validation datasets. Newly identified COVID-19 cases are reported across time and are broken down by HOCI and COCI case types. Panel A shows the non-UK (Geneva) hospital case load during an epidemic surge of cases. Panel B shows the UK hospital group post pandemic surges 1 and 2, when COVID-19 became endemic and non-surging.
The results showed that dynamic patient contact networks could be a strong predictor of transmission of respiratory viral infections in hospitals. Thus, the integration of these networks in clinical care could improve individualized prevention of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection as well as early diagnosis.
“The framework provides daily patient-individualized predictions and can identify dynamic disease acquisition risk factors.”
They found that centrality measures calculated from patient contact networks together with some hospital contextual variables represent a key HOCI risk factor, which can boost the performance of predictive models in a pandemic as well as endemic conditions.
To demonstrate the transferability of the framework, they applied it to data from a Geneva-based hospital during COVID-19 surge 1 and showed increased predictive power by including contact network risk factors. However, access was only available for room-level contacts that were less predictive in the London data.
With no end in sight to the COVID-19 pandemic, this new predictive framework and contact risk factors offer a valuable tool to assist identification of potential HOCI cases across various epidemic and endemic periods.
“Further work will be needed to extend this work to other healthcare-acquired infections by assessing how the inclusion of variables capturing a patient’s environment within an underlying contact network could aid prediction with a view to informing infection prevention and control measures.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Predicting hospital-onset COVID-19 infections using dynamic networks of patient contacts: an observational study Ashleigh Myall, James R Price, Robert L Peach, Mohamed Abbas, Siddharth Mookerjee, Nina Zhu, Isa Ahmad, Damien Ming, Farzan Ramzan, Daniel Teixeira, Christophe Graf, Andrea Y Weiße, Stephan Harbarth, Alison Holmes, Mauricio Barahona medRxiv 2021.09.28.21264240; doi: https://doi.org/10.1101/2021.09.28.21264240, https://www.medrxiv.org/content/10.1101/2021.09.28.21264240v1
- Peer reviewed and published scientific report.
Myall, Ashleigh, James R. Price, Robert L. Peach, Mohamed Abbas, Sid Mookerjee, Nina Zhu, Isa Ahmad, et al. 2022. “Prediction of Hospital-Onset COVID-19 Infections Using Dynamic Networks of Patient Contact: An International Retrospective Cohort Study.” The Lancet Digital Health 4 (8): e573–83. https://doi.org/10.1016/S2589-7500(22)00093-0. https://www.thelancet.com/journals/landig/article/PIIS2589-7500(22)00093-0/fulltext.