The coronavirus disease 2019 (COVID-19), which is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has claimed over 4.8 million lives worldwide and continues to be a major global threat.
The detection of early prognostic markers of COVID-19 is of the utmost importance as a result of the high mortality and severity of the disease. Poor outcomes related to COVID-19 are not only due to viral infection but also due to aberrant host immune response.
 Study: Nasopharyngeal Microbiota as an early severity biomarker in COVID-19 hospitalised patients: a retrospective cohort study in a Mediterranean area. Image Credit: qcontrol / Shutterstock.com
Study: Nasopharyngeal Microbiota as an early severity biomarker in COVID-19 hospitalised patients: a retrospective cohort study in a Mediterranean area. Image Credit: qcontrol / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Microbiota and COVID-19
The severity of COVID-19 has been related to several prognostic factors such as age, comorbidities, sex, genetic factors, and geographical locations. These characteristics help to promote immune responses, as well as prevent excessive antiviral immune responses.
The microbiota is known to influence the natural history of many infectious diseases. The clinical evolution of chronic respiratory diseases, as well as viral respiratory infections, has also been correlated with respiratory microbiota. Therefore, several studies involving COVID-19 pathology and microbiota have been conducted to determine the role of microbiota in an individual’s susceptibility to SARS-CoV-2 infection.
Several of these studies have confirmed the relationship between the composition of an individual’s respiratory and gut microbiota and disease severity. Despite these findings, longitudinal studies evaluating microbiota as a prognostic factor for disease severity are still lacking.
A new study published on the preprint server bioRxiv* aimed to analyze the nasopharyngeal microbiota of patients who were hospitalized with COVID-19. Here, the researchers also were also interested in determining the relationship between the SARS-CoV-2 clinical outcomes and the microbiota by identifying the genera or features that could serve as prognostic markers.
About the study
The current study involved a retrospective cohort of 177 adult patients who were hospitalized with COVID-19 between February 27th, 2020 to January 22nd, 2021. The SARS-CoV-2 infection was confirmed through the reverse-transcriptase polymerase chain reaction (RT-PCR) test.
Upon admission to the hospital, nasopharyngeal samples were collected and stored at -80 °C. These samples were later used for deoxyribonucleic acid (DNA) isolation and quantification.
The microbial diversity measured by the alpha and beta diversity indexes, as well as the taxonomic composition of the microbiota, was determined by bioinformatic analysis and microbiota amplicon next-generation sequencing (NGS).
The clinical features, laboratory and radiological tests, presence of comorbidities, prescribed therapies, and outcomes during the acute phase of SARS-CoV-2 infection of patients were collected from their digital medical records.
Study findings
Out of the 177 patients included in the current study, 57.6% were males and there was a median age of 68 years. Assessment of the patients after six days of symptoms showed that 89.2% had pneumonia. The mortality rate was found to be 17. 5%, with 11.3% of the patients requiring invasive mechanical ventilation (IMV).
The alpha and beta diversity indexes were found to be lower in patients with fatal outcomes, thus further suggesting that low microbial diversity is associated with higher COVID-19 severity.
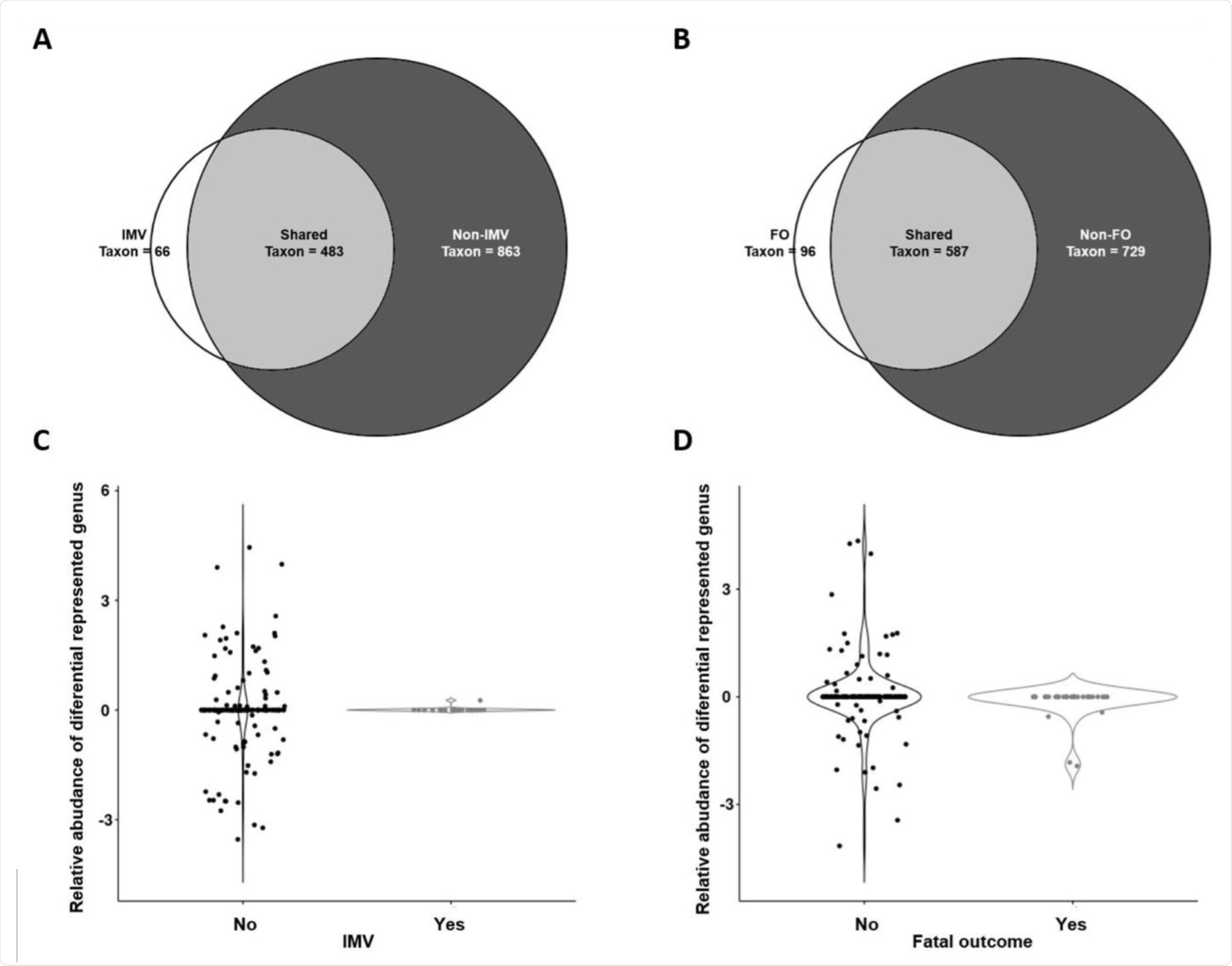 Taxonomic analysis: Venn diagrams for IMV (A), and fatal outcome (B), and relative abundances of differential genera for IMV (C), and fatal outcome subpopulations (D). Relative abundances are shown in logarithmic scale. IMV: invasive mechanical ventilation, FO: Fatal outcome.
Taxonomic analysis: Venn diagrams for IMV (A), and fatal outcome (B), and relative abundances of differential genera for IMV (C), and fatal outcome subpopulations (D). Relative abundances are shown in logarithmic scale. IMV: invasive mechanical ventilation, FO: Fatal outcome.
Furthermore, few genera of bacteria were found to be more abundant in COVID-19 patients, irrespective of patients belonging to the IMV or fatal outcome groups. These bacteria included Streptococcus spp., Staphylococcus spp., and Corynebacterium spp.
The researchers also found that Selenomonas spp., Filifactor spp., Chroococcidiopsis spp., and Actinobacillus spp. were more abundant in non-IMV patients. Comparatively, the presence of Selenomonas spp., Filifactor spp., Actinobacillus spp., or Chroococcidiopsis spp were associated with a reduced risk of IMV.
In patients who succumbed to fatal outcomes, Actinobacillus spp., Craurococcus spp., Citrobacter spp., and Moheibacter spp. were found to be more abundant in non-exitus patients. Actinobacillus spp., Citrobacter spp., Craurococcus spp., or Moheibacter spp. were associated with a reduced risk of fatal outcomes. However, this relationship did not persist after adjustment for the main confounders.
The study is clinically relevant, as it was low cost and allowed for the rapid detection of the diversity and genera of microbes during the admission of patients to the hospital. Furthermore, the current study was effective in determining that higher diversity was found in patients without fatal outcomes or IMV. The current study also showed that certain specific genera of microbes were present in patients hospitalized with COVID-19 that could serve as early biomarkers.
However, the relationship between microbiota and COVID-19 is still an active and expanding field of research. Future research needs to be done to determine their exact roles in COVID-19 development and evolution.
Limitations
The current study was an observational, single-center, retrospective study where the collection of data was not standardized. The small sample size, along with the absence of differences in characteristics among the patients admitted to the hospital, did not allow for global assessment.
The 16S ribosomal RNA amplicon sequencing approach could also introduce bias. Moreover, the bioinformatics analysis of microbes has not been standardized yet, which could hamper the interpretation of the results.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Ventero, M. P., Moreno-Perez, O., Molina-Pardines, C., et al. (2021). Nasopharyngeal Microbiota as an early severity biomarker in COVID-19 hospitalised patients: a retrospective cohort study in a Mediterranean area. bioRxiv. doi:10.1101/2021.09.28.461924. https://www.biorxiv.org/content/10.1101/2021.09.28.461924v1
- Peer reviewed and published scientific report.
Ventero, Maria Paz, Oscar Moreno-Perez, Carmen Molina-Pardines, Andreu Paytuví-Gallart, Vicente Boix, Isabel Escribano, Irene Galan, et al. 2022. “Nasopharyngeal Microbiota as an Early Severity Biomarker in COVID-19 Hospitalised Patients.” Journal of Infection 84 (3): 329–36. https://doi.org/10.1016/j.jinf.2021.12.030. https://www.journalofinfection.com/article/S0163-4453(21)00647-2/fulltext.