There are four types of coronaviruses (CoVs), of which include Alpha, Beta, Gamma, and Delta coronavirus. Among them, Beta CoVs are positive-stranded, enveloped ribonucleic acid (RNA) viruses that affect a wide range of hosts. The human-specific species of Beta CoVs are capable of causing diseases associated with a range of effects including mild respiratory symptoms to life-threatening forms of atypical pneumonia like the severe acute respiratory syndrome (SARS).
The envelope of CoVs is made up of three structural proteins that include the spike (S), membrane (M), and envelope (E) proteins, as well as certain accessory proteins. The S protein is essential for the attachment of the virus to the host cell angiotensin-converting enzyme 2 (ACE2) receptor for SARS-CoV-2 entry, as well as the fusion between the viral and target cell membrane. The E and M proteins are mostly required for the viral assembly.
All the three structural proteins of CoVs are synthesized in the endoplasmic reticulum (ER). Furthermore, these proteins may also undergo post-translational modifications, such as S-acylation on their cytosolic parts and N-glycosylation on their luminal parts. S-acylation, which is the covalent attachment of medium-chain fatty acids to the S atoms of cytosolic cysteines, is a common process that occurs in mammalian cells and is also a hallmark of viral envelopes.
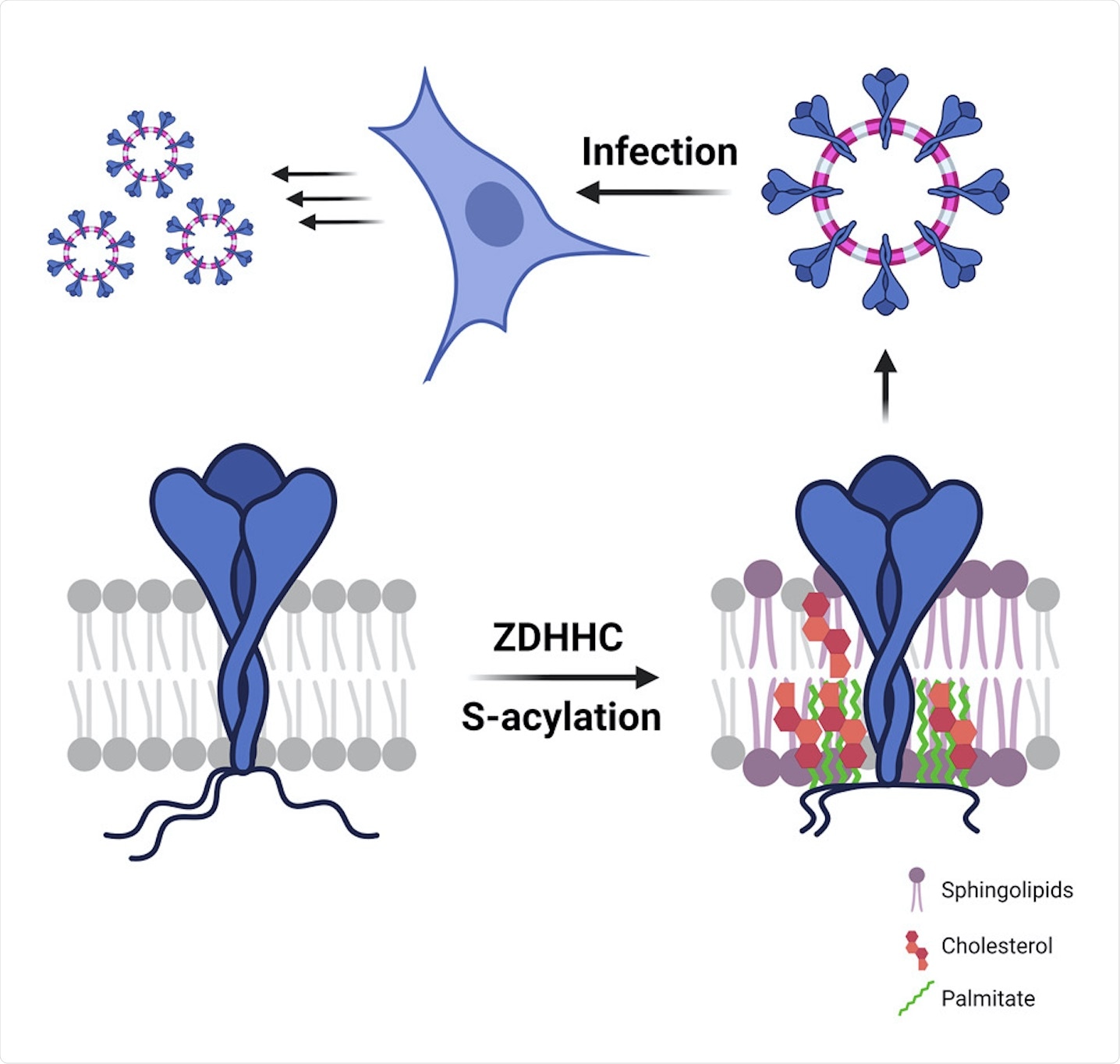 Study: S-acylation controls SARS-CoV-2 membrane lipid organization and enhances infectivity.
Study: S-acylation controls SARS-CoV-2 membrane lipid organization and enhances infectivity.
A new study published in Developmental Cell investigates S-acylation of the SARS-CoV-2 S protein. To this end, the researchers of this study aimed to identify the host enzymes that are involved in the process, as well as determine the importance of this acylation for viral biogenesis and infection.
How are SARS-CoV-2 viral proteins acylated?
Similar to the findings from SARS-CoV-1, the S protein from SARS-CoV-2 also undergoes S-acylation when expressed in monkey kidney Vero E6 cells or Hela cells. The current study involved the SARS-CoV-2 D614G variant that emerged during the early stages of the pandemic and spread throughout the world.
Two assays were used to monitor the protein acylation, which included the acyl-resin assisted capture assay (Acylrac) and autoradiographic assay. The Acylrac assay involved the capture of the S-acylated proteins with the help of thiol-reactive sepharose beads. Comparatively, the second assay involved the incubation of cells with radioactive H3-palmitate followed by monitoring its incorporation into the S protein with the help of immunoprecipitation and autoradiographic analysis. Both of these assays helped in the detection of acylation of all the S protein fragments.
S-acylation is carried out by a family of 23 protein acyltransferases that are defined by the presence of a conserved zinc-finger domain with a DHHC motif (ZDHHCs). For identification of the enzyme that modified the S protein, all ZDHHCs were depleted in Hela cells. ZDHHC8, 20, and 9 were found to play major roles in S protein acylation.
Next, the involvement of these identified ZDHHCs during infection was tested in Vero E6 cells. It was found that the depletion of ZDHHC8 also led to a reduction in ZDHHC20 messenger RNA (mRNA) in monkeys, while no such effect was seen in human cell lines. Furthermore, knocking down ZDHHC9 and 20 led to a decrease in S protein S-acylation during infection.
With the help of Vero E6 cells, the researchers found that knocking out of one transferase did not affect the role of the others. Finally, it was concluded that ZDHHC20 played a major role in S protein modification during infection followed by ZDHHC9.
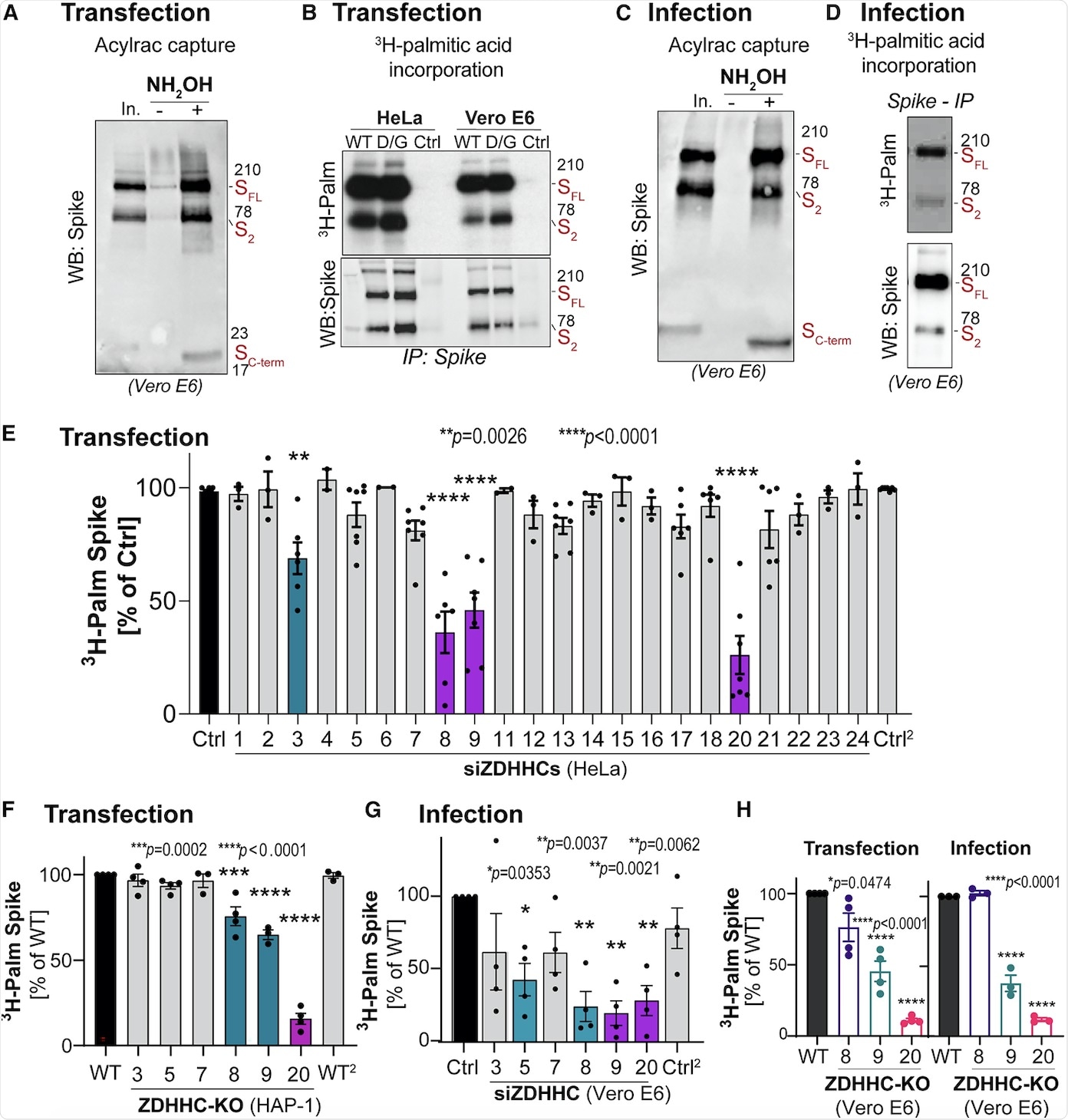 Identification of the spike-modifying ZDHHC acyltransferases.
Identification of the spike-modifying ZDHHC acyltransferases.
Acylation during S protein biogenesis
S proteins are known to contain a conserved cysteine-rich stretch of amino acids in their cytoplasmic tails. For SARS-CoV-2, the number of cysteine residues is 10 within the first 20 amino acids.
The current study generated four HA-tagged S protein mutants, where 2-3 cysteine amino acids (group I) were changed to alanine. This led to an 80% reduction in H3-palmitate incorporation.
Mutation of the next three cysteines (group II) to alanine also led to a 40% reduction in H3-palmitate incorporation. A combination of group I and II had an effect that was equivalent to changing all the 10 amino acids. From this analysis, it could be concluded that all the 10 cysteines were targeted for S-acylation.
Following this, analysis of H3-palmitate incorporation into HA-tagged S protein mutants in Hela cells, a comparison between H3-palmitate incorporation into the S protein in Hela cell transfection and Vero E6 cells infection was conducted. The results of this experiment indicated that for the first case, lower levels of incorporation occurred, while in the second case the level of incorporation was identical in both transfection and infection.
An investigation of where the S protein acylation occurs during viral biogenesis was also conducted. The S protein trimers are transported to the ER-Golgi intermediate after synthesis, folding, and assembly. Determination of whether the S protein acylation took place during protein biogenesis was monitored with the help of protein synthesis inhibitor cycloheximide (CHX) and the Golgi-disrupting drug brefeldin A (BrfA) in HA-tagged S protein variants.
Palmitate incorporation was found to decrease by approximately 50% when treated with either CHX or BrfA. This suggested that S-palmitoylation was initiated shortly after protein synthesis and continued after the protein exited the ER.
The kinetics of H3-palmitate incorporation was also monitored when the S protein-modifying ZDHHC enzymes were silenced. These results indicated that silencing of either ZDHHC20, ZDHHC8, or ZDHHC9 had drastic effects, whereas the silencing of ZDHHC20 had the earliest and most dramatic effect.
Labeling of S protein cysteines with maleimide-PEG also showed that the entire S protein population produced by the infected cells undergoes massive and rapid acylation on all the 10 cysteine residues.
S protein acylation is stable during infection
S-acylation could be reversed by deacetylating enzymes such as acyl protein thioesterases (APTs). H3-palmitate decay analysis was done in transfected Hela cells that showed a reduction in palmitoylation of the S protein; however, the same analysis yielded different results during SARS-CoV-2 infection in Vero E6 cells. It was found that during infection in Vero E6 cells, no loss of palmitate from the S protein occurred while depalmitoylation of other cellular proteins was detected.
Effect of S-acylation on the biogenesis of S protein and its turnover rate
The researchers then conducted two separate experiments to determine whether S-acylation modified the turnover of the S protein. The first was a 35S Cys/Met metabolic pulse-chase experiment, which helped to monitor the evolution of a protein starting from its synthesis, and cycloheximide chase, which helped to monitor the degradation of a fully folded protein.
The results of the Cys/Met metabolic pulse-chase experiment indicated that acylation by ZDHHC20 led to a 3-fold increase in the total amount of the S protein. The cycloheximide chase, in turn, determined that S-acylation slowed down the turnover of the S protein, thereby increasing its turnover from 2 hours to 4 hours.
How does S protein S-acylation modify the organization of the lipid bilayer?
The current study also discussed the effect of acylation on lipid organization, for which comparison of palmitoylated and non-palmitoylated S proteins took place in a model membrane. The composition of the model membrane was as follows: 50% dipalmitoylphosphatidylcholine (DPPC), 30% dilinoleylphosphatidylcholine (DLiPC), and 20% cholesterol (CHOL).
The results indicated that acylation led to the collapse of the cytoplasmic tails that helped to bring the palmitate moieties close to the transmembrane, which further led to a 9-fold increase in local acyl chain concentration. Furthermore, an unmodified S protein was associated with the disordered phase, whereas an acylated S protein was associated with the ordered phase. Therefore, it could be concluded that an acylated S protein could drive trans-bilayer lipid asymmetry.
How does S-acylation of S protein drive its association with ordered lipid nanodomains?
For the determination of the lipid environment of the S protein, the researchers tested whether transfected intracellular S proteins could be found in detergent-resistant membranes (DRMs). DRMs have been considered as important markers to determine the association between proteins and cholesterol-rich lipid domains. The results indicated that the intracellular S protein was only found in DRM fractions, while the cell surface S protein was distributed throughout the gradient.
The DRM association of the S protein was lost when treated with ZDHHC20 small interfering RNA (siRNA), while silencing of ZDHHC9 did not show these results. Thus, it could be concluded that ZDHHC20-mediated acylation of the S protein led to its association with DRMs.
How does S-acylation of S protein control the local lipid environment in viral-like particles?
The current study also investigated whether the S-acylation of the S protein altered its lipid environment. This investigation was done with the help of two viral particle assembly systems including an HIV-derived lentivector-based system to generate S protein harboring pseudo particles (PPs) and a SARS-CoV-based system to generate viral-like particles (VLPs). The results indicated that the presence of an acylated S protein could alter the organization of its local lipid environment.
Unusual lipid composition of SARS-CoV-2 virions
The current study also included the biochemical analysis of virions purified from infected Vero E6 cells that were silenced for acyltransferase expression using MixT. Similar levels of S and M proteins were observed in cells and virions, irrespective of acylation. Cryo-electron microscopy (cryo-EM) suggested that the ZDHHC acyltransferases 9, 8, and 20 led to tight control of viral particle dimensions.
Furthermore, the DRM association of the S protein within the SARS-CoV-2 envelope was analyzed. The S protein was found in DRMs, only in the presence of acylation.
The lipid composition of SARS-CoV-2 virions showed that they were poor in glycerolipids, enriched in sphingolipids, and deprived of non-membrane lipids. Silencing of the acyltransferases, however, led to a 30% reduction in cholesterol amounts in the virion.
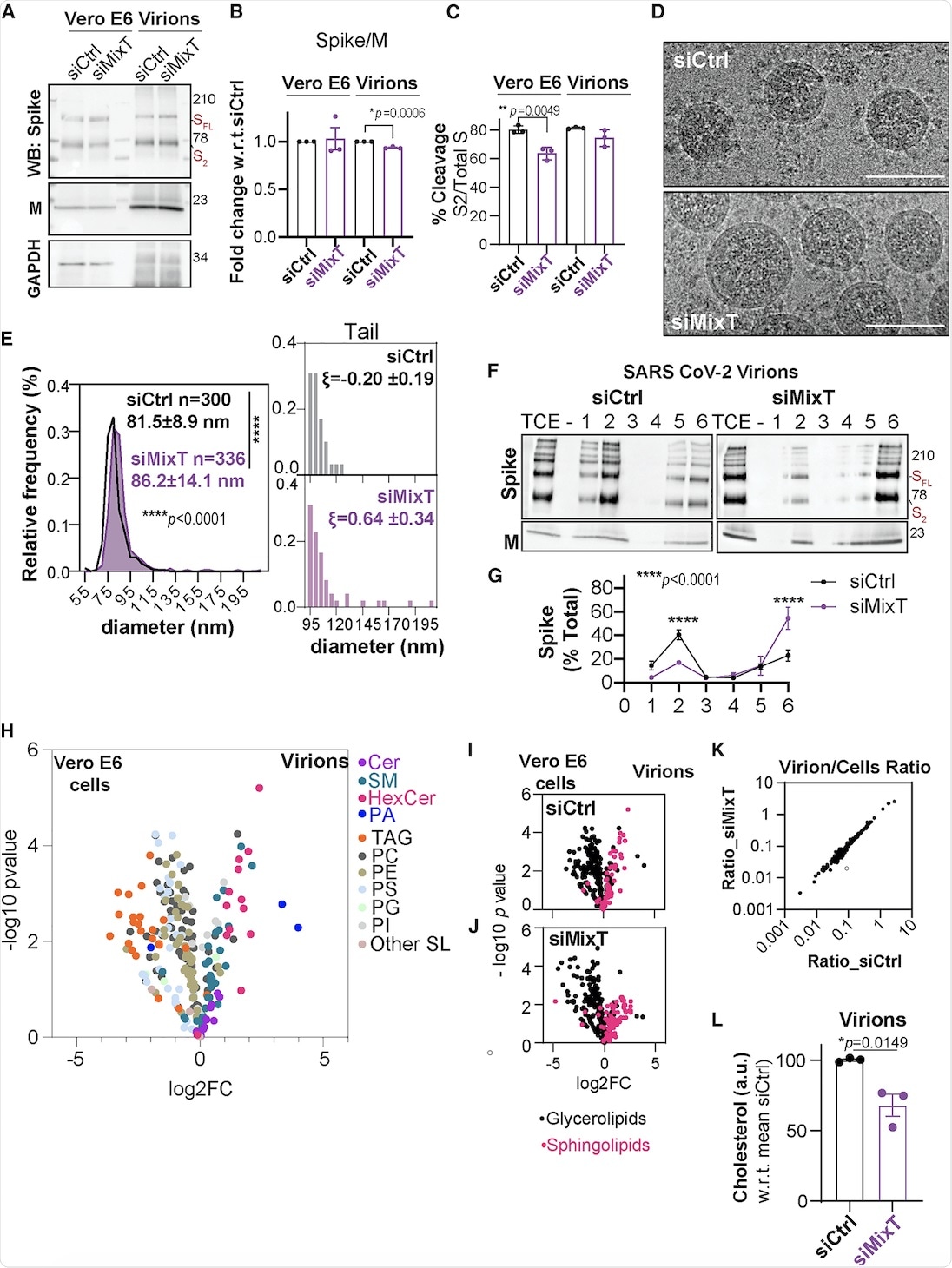 Impact of S-acylation on SARS-CoV-2 virion biogenesis.
Impact of S-acylation on SARS-CoV-2 virion biogenesis.
Importance of S-acylation for SARS-CoV-2 fusion with the host cell
Finally, an investigation of the role of the S protein S-acylation in SARS-CoV-2 virion infection took place. At first, the competence of ZDHHC20 and ZDHHC9 KO cells for SARS-CoV-2 infection was analyzed. This was followed by an analysis of the infectivity of the viruses produced by ZDHHC-KO cells. It was found that the infectivity of the virions produced from either ZDHHC20 and ZDHHC9 KO cells was 30 to 40% less as compared to the control virions.
Furthermore, fungal metabolite myriocin was used to prevent the formation of cholesterol and sphingolipid-rich domains that could affect SARS-CoV-2 infection. The results showed that the disruption of the sphingolipid cascades led to inhibition of S-acylation during infection.
Additionally, assessment of the role of S protein acylation took place with the help of two viral particle systems, PPs and SARS-CoV VLPs. For both PPs and VLPs, it was found that the absence of S protein acylation led to reduced fusion with the host membrane and, as a result, reduced entry into the host cell.
Conclusion
The current study is based on a combination of functional genomics, biochemical, kinetic, and computational studies. The results presented here help to determine S protein acylation that occurs during SARS-CoV-2 biogenesis, as well as the importance of this acylation.
The results of the study indicate that S-acylation could be a promising drug target for CoV infection. Previous studies with influenza viruses have already shown ZDHHC20 to be involved in acylation.
Recent structural elucidation of this enzyme makes it specifically druggable. Thus, further research needs to be carried out regarding S-acylation, as it is not only an important regulatory mechanism for CoVs, but also all enveloped viruses in general.