Researchers in Germany have developed a promising approach to suppressing the viral replication of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – the agent that causes coronavirus disease 2019 (COVID-19).
The team – from the Technical University of Berlin, the Free University of Berlin and the German Center for Infection Research in Charitéplatz, Berlin – designed eight small interfering RNAs (siRNAs) that target a highly conserved region of SARS-CoV-2.
As reported in the journal Viruses, the most efficient of these siRNAs – siCoV6 – significantly inhibited the replication of SARS-CoV-2 virions and was active against the B.1.1.7 (alpha) variant.
Interestingly, siCoV6 was even highly active against a member of the SARS-CoV-1 family, pointing to a potent siRNA with broad antiviral activity against SARS-CoV viruses.
Jens Kurreck and colleagues say this broad antiviral activity makes siCoV6 a promising candidate for the treatment of SARS coronavirus infections.
The development of effective antivirals is urgently needed
Since the COVID-19 outbreak began in late December 2019, the causative agent SARS-CoV-2 has infected more than 238 million people and claimed the lives of more than 4.86 million.
Despite the successful development and approval of several vaccines, treatment options for patients with severe disease are limited and effective antiviral drugs are urgently needed.
The SARS-CoV-2 RNA genome is approximately 30 kilobases in length and codes for 16 non‐structural proteins (nsp1–16) that are required for RNA replication.
Once SARS-CoV-2 has entered a host cell, all of these proteins are directly translated and induce replication of the RNA genome.
A unique feature of coronaviruses is that not only the complete genomic RNA, but also subgenomic RNAs are produced. These subgenomic RNAs all share the same 5’‐end (of around 75 nucleotides), which is referred to as the “leader sequence.”
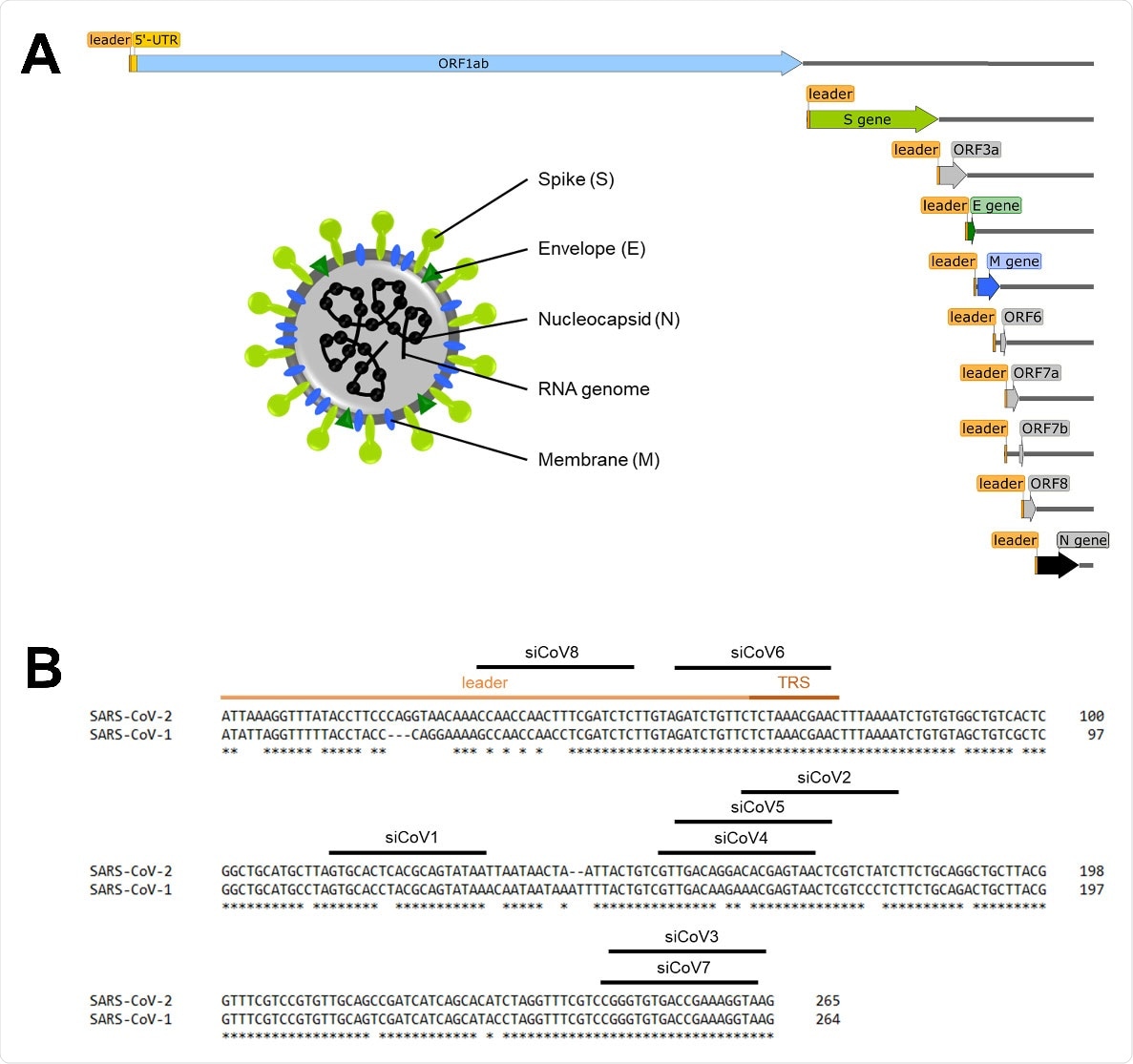
Schematic overview of the SARS‐CoV‐2 genome and target sequences of the siRNAs in the 5’‐UTR. (A) Schematic overview of the SARS‐CoV‐2 genome, the subgenomic RNAs and the virion structure. The coronavirus virion consists of the structural spike protein (S), envelope protein (E), membrane protein (M) and nucleoprotein (N). The single‐stranded RNA genome with positive polarity is encapsulated by N, while S‐trimers protrude from the host‐derived virus envelope and enable binding to new host cells. In addition to the genomic RNA, nine subgenomic RNAs are synthesized during replication, all of which share the same 5’‐ and 3’‐ends. The orange box represents the mutual 5’‐end, the leader sequence. (B) Target sequences of the siRNAs in the 5’‐UTR of SARS‐CoV‐2. The leader sequence (light orange) and the transcription regulatory sequence (TRS, dark orange) can be found at the 5’‐end of the genomic and subgenomic RNAs. Nucleotides without an asterisk differ from the SARS‐CoV‐1 5’‐UTR sequence.
A promising approach to inhibiting viral replication
A promising approach to inhibiting viral replication is the use of RNA interference (RNAi) – an efficient post‐transcriptional gene silencing process that can be triggered by siRNAs. This process leads to the degradation of messenger RNA (mRNA) in a sequence‐specific manner and inhibits gene expression.
The targeting of gene expression through RNAi is being developed as a new therapeutic strategy, and numerous RNAi‐based agents have previously been investigated to treat viral infections. RNA viruses such as SARS‐CoV‐2 are particularly promising candidates since not only their mRNAs but also their genomic RNA can be directly targeted.
In line with this, Kurreck and colleagues and other research groups have previously shown that the replication of RNA viruses such as influenza and hepatitis C can be effectively inhibited by RNAi in various cell and animal disease models.
What did the current study involve?
The team designed and tested eight siRNAs directed against the 5’-untranslated region (5’-UTR) of SARS-CoV-2.
The 5’‐UTR is a highly conserved region that is crucial for viral RNA replication and transcription, making it a promising target for the design of RNAi‐based treatments.
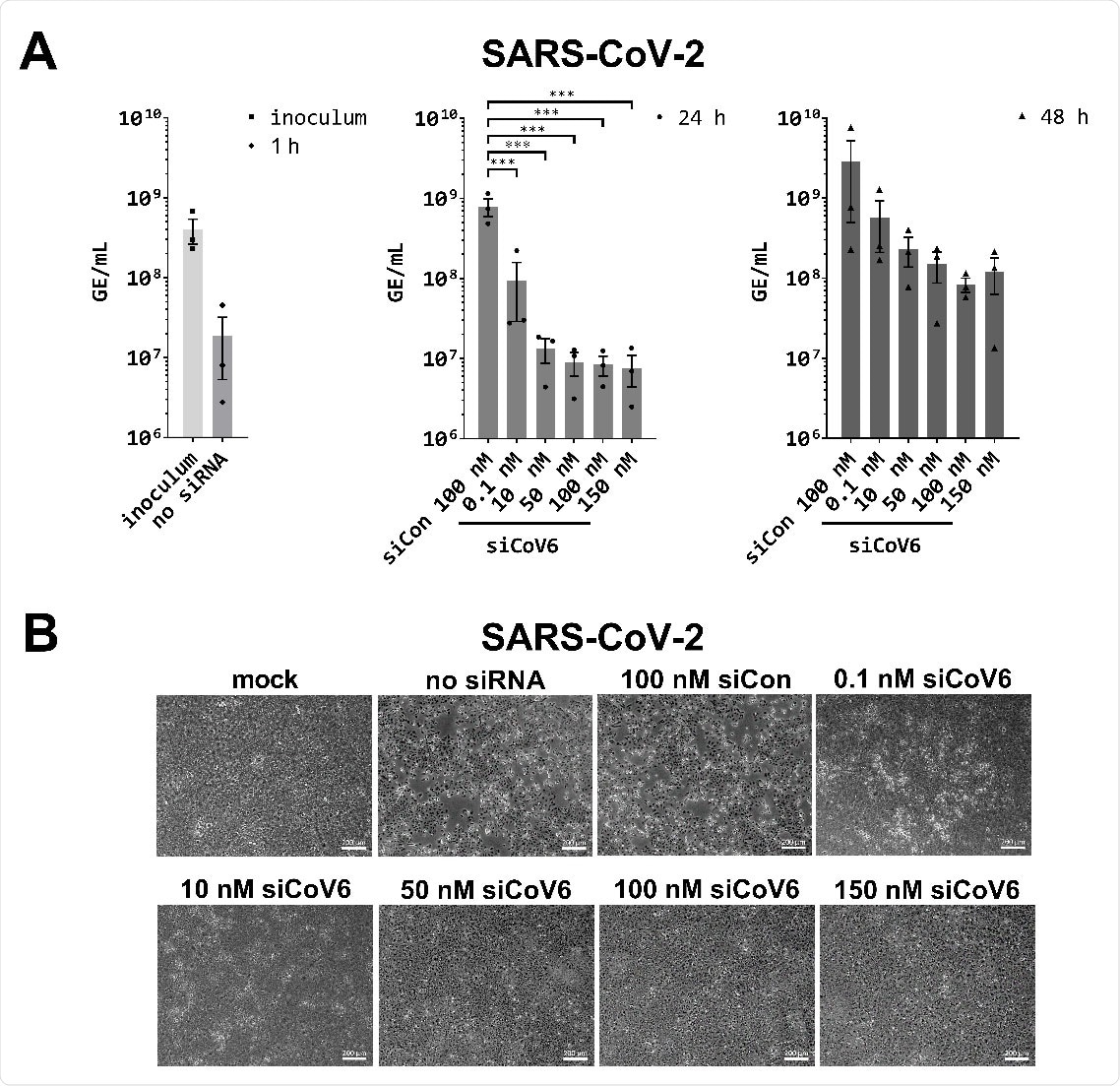
Inhibition of SARS‐CoV‐2 replication by siCoV6. (A) The replication of SARS‐CoV‐2 (BetaCoV/Munich/BavPat2‐ChVir984‐ChVir1017/2020) was investigated by quantitative RT‐PCR in Vero E6 cells. Vero E6 cells were transfected with 0.1–150 nM siCoV6 or 100 nM siCon and infected 24 h after transfection with SARS‐CoV‐2 at a MOI of 0.01. Subsequently, the cells were washed, supplemented media were added and viral RNA was isolated from the culture supernatant at 1 hpi, 24 hpi and 48 hpi. Genome equivalents per mL (GE/mL) were determined by quantitative RT‐PCR. The mean ± SEM of three independent experiments is shown. The statistical significance was determined by a univariate analysis of variance (one‐way ANOVA); ***p <0.001. (B) Cell morphology observation. Cells were fixed 48 h after infection with SARS‐CoV‐2 and the cytopathic effect (CPE) of viral infection was documented by transmitted light microscopy. Representative data from three independent experiments are shown. Scale bar: 200 μm.
Initial reporter assays confirmed that all eight siRNAs efficiently targeted the 5’‐UTR of SARS‐CoV‐2.
The most promising candidate in the panel – siCoV6 – targeted the leader sequence that is present in both the viral genome and all the subgenomic RNAs that are essential for replication of the virus.
When tested with infectious SARS-CoV-2, siCoV6 effectively inhibited virus replication in Vero E6 cells by two orders of magnitude (99%) at only 10 nM siRNA and protected cells from cytopathic effect.
Furthermore, since its target sequence is highly conserved, siCoV6 also showed promising inhibitory activity against the alpha variant of SARS‐CoV‐2 and even against a member of the SARS‐CoV‐1 family.
Kurreck and colleagues say the results clearly showed that siCoV6 inhibits the replication of SARS‐CoV‐2 and protects cells from infection‐induced cell toxicity.
Further analyzing the effects of siCoV6 on variants
Since SARS‐CoV‐2 is constantly evolving and mutations can result in the alteration of siRNA target sequences, the team analyzed around 1500 of the most recent sequences of different SARS‐CoV‐2 variants with respect to mutations in the target site of siCoV6.
The researchers did not identify any mutations in the target site of the B.1.351 (beta) or B.1.617.2 (delta) variant, and for the alpha and P.1 (gamma) variants, only less than 0.8% of sequences had mutations in the siCoV6 target site.
“This bioinformatic analysis confirms the eligibility of the 5’‐UTR and in particular the leader sequence as a target site for the implementation of RNAi as a broad‐range antiviral approach,” writes the team.
Kurreck and colleagues say this work has identified a very potent siRNA with a broad activity against various SARS-CoV viruses that represents a promising candidate for the development of new COVID-19 treatment options.