Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) invades host cells and, using cellular resources, rapidly proliferates. To control the coronavirus disease 2019 (COVID-19) pandemic and decipher the virus's molecular evolution, it is essential to understand how SARS-CoV-2 utilizes host cellular mechanisms to thrive. There have been many efforts to understand the molecular mechanisms of SARS-CoV-2 better. However, a definitive explanation of why this virus is so well adapted to invade human cells remains lacking.
.jpg) Study: SARS-CoV-2 competes with host mRNAs for efficient translation by maintaining the mutations favorable for translation initiation. Image Credit: joshimerbin/ Shutterstock
Study: SARS-CoV-2 competes with host mRNAs for efficient translation by maintaining the mutations favorable for translation initiation. Image Credit: joshimerbin/ Shutterstock
A successful human cell invasion for SARS-CoV-2 is measured by how many viral particles are produced. Fast translation of viral RNAs is the basis of the rapid production of viral proteins, which requires further binding and scanning of ribosomes at the 5-prime end of mRNA, which allows for efficient translation initiation. Several approaches (cis and trans) are utilized by hosts that enhance the translation initiation efficiency, such as improving the initiation factors to elevate their efficiency or changing ribosomes to types with higher affinity to mRNAs.
A study published in the Journal of Applied Genetics shows that mutations that SARS-CoV-2 has gained improve its advantage in the efficient translation of viral RNAs. The authors explain that this strategy may assist the virus in adapting to human host cells.
The study
SARS-CoV-2 has 11 non-redundant coding sequences CDSs. The authors divided each CDS into ten bins with equal length and then calculated each bin's GC content. A significant difference was found in the SARS-CoV-2 5-prime bin, which had a low GC content compared to the other CDS. Human genes were found to have high GC content at the 5-prime end of CDS compared with the other parts. Due to only 11 genes being available with SARS-CoV-2, the was much more variance displayed. Opposite patterns between SARS-CoV-2 and humans were observed in the global trend.
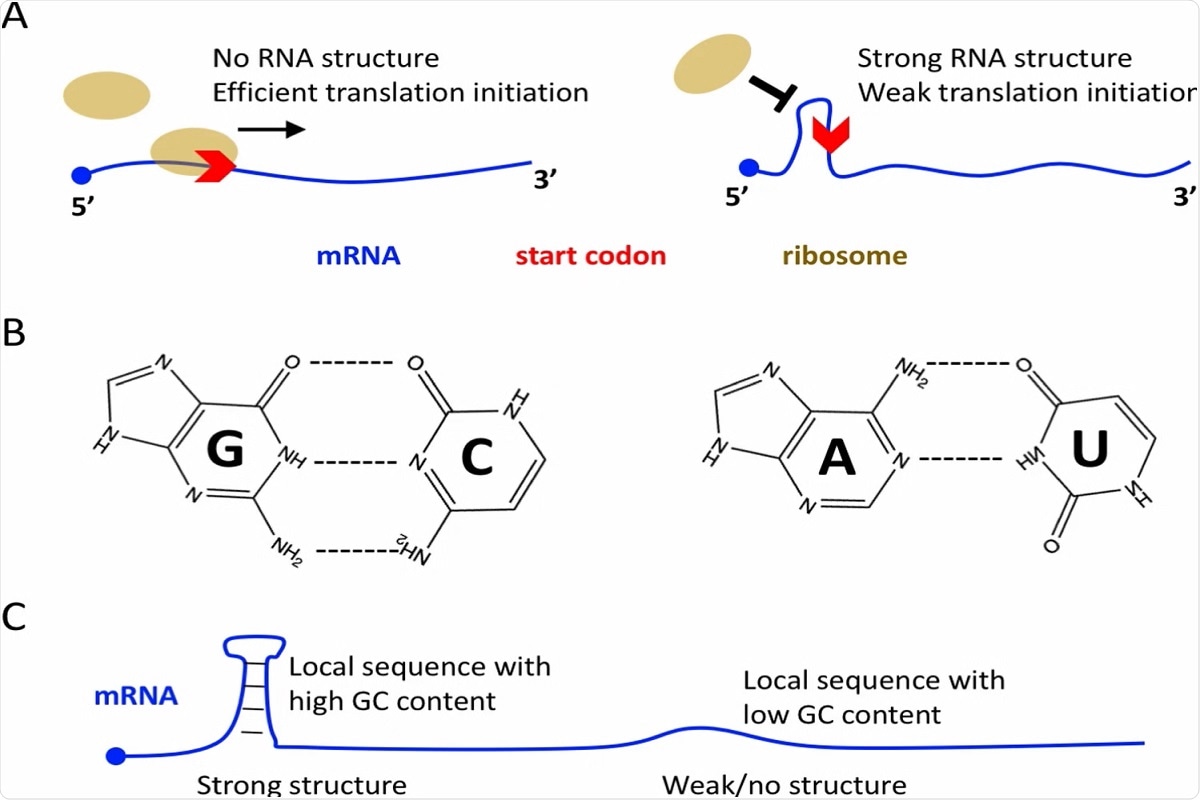 Molecular basis of mRNA translation initiation. A No RNA structure near the start codon is favorable for efficient translation initiation. Strong RNA structure near the start codon usually leads to low initiation efficiency. B G:C base pair is biochemically more stable than A:U base pair. C Local RNA sequence with high GC content will lead to strong local structure, and vice versa
Molecular basis of mRNA translation initiation. A No RNA structure near the start codon is favorable for efficient translation initiation. Strong RNA structure near the start codon usually leads to low initiation efficiency. B G:C base pair is biochemically more stable than A:U base pair. C Local RNA sequence with high GC content will lead to strong local structure, and vice versa
RNA structure stability is likely to increase with mutations resulting in higher GC content, such as A>G and T>C mutations. In contrast, an RNA structure could be unravelled or loosened by mutations that decrease GC content, such as G>A and C>T mutations.
The authors observed a decrease in RNA structure at the 5-prime CDS of SARS-CoV-2, so if this mechanism was advantageous to SARS-CoV-2, the virus sequence should continue to optimize. The mutation profile could reveal this long-lasting optimization process.
Fixed and polymorphic mutations are classifications that could be applied to these mutations. For example, within SARS-CoV-2, the fixed mutations are those diverged sites between RaTG13 and SARS-CoV-2, representing a relatively long timescale. Among the worldwide SARS-CoV-2 strains, the polymorphic mutations were detected in variant sites, which are now shaping natural selection events.
Between host cells and viruses, evolution is an ongoing arms race. Viral sequence "upgrading" does not guarantee successful viral invasion because optimization of host mRNA can also occur. SARS-CoV-2 mutations had a global trend to elevate the translation initiation efficiency of viral RNAs, while human mutations were "neutral" regarding host mRNA translation.
Conclusion
SARS-CoV-2 is, for the moment, ahead in the evolutionary arms race in terms of competing for ribosomes and rapid translation. However, this is not implying that novel mutations have any biases in SARS-CoV-2, as novel mutations are random. Still, viruses replicate faster than host cells, which allows for the accumulation of more mutations. Unfavorable mutations in viral sequences are eliminated by natural selection, leaving only the advantageous ones.