Researchers in the United States have developed a cost-effective, high-throughput pipeline for identifying effective nanobodies that target and neutralize severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – the agent that causes coronavirus disease 2019 (COVID-19).
The most potent nanobody the team identified – RBD-1-2G – tolerates the N501Y mutation found in the receptor-binding domain (RBD) of the surface spike protein of several SARS-CoV-2 variants.
The spike protein mediates the initial stage of the infection process when its RBD binds to the host cell receptor angiotensin-converting enzyme 2 (ACE2). The spike RBD is a primary target of antibodies following vaccination or natural infection.
Anton Simeonov from the National Institutes of Health in Rockville, Maryland, and colleagues found that the potent RBD-1-2G nanobody was capable of neutralizing pseudotyped virus particles containing N501Y.
Furthermore, a primary human tissue model demonstrated that RBD-1-2G-Fc treatment reduced viral burden following infection with the B.1.1.7 (alpha) variant of SARS-CoV-2.
The method described here could also be expanded to other infectious diseases, says the team.
A pre-print version of the research paper is available on the bioRxiv* server, while the article undergoes peer review.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The need for effective therapeutics is ongoing
Since the COVID-19 outbreak began in late December 2019, an unprecedented response among the scientific community quickly resulted in the development of effective vaccines against the causative agent SARS-CoV-2.
Although these vaccines have been shown to significantly reduce the incidence of severe disease and hospitalizations, the emergence of novel SARS-CoV-2 variants exhibiting resistance to vaccine induced-immunity has raised concerns about ongoing transmission.
The need for high-throughput, adaptable pipelines capable of identifying effective therapeutics against SARS-CoV-2 is therefore ongoing.
More about the spike protein and the RBD
The SARS-CoV-2 infection process is mediated by the viral spike glycoprotein, which recognizes host ACE2 receptors via its RBD and contains the fusion machinery needed for viral entry.
Mutations in this spike have been implicated in increased viral infectivity and the RBD is the focus of intense interest for the development of neutralizing antibodies.
Multiple variants of SARS-CoV-2 have arisen in which N501 – a key contact residue within the RBD – has been mutated to tyrosine (N501Y). This mutation increases the spike’s binding affinity for ACE2.
Where do nanobodies come in?
Structural insights into how antibodies bind to SARS-Cov-2 variants are still needed and anti-spike-RBD neutralizing antibodies have been isolated from different platforms.
Nanobodies are a unique class of heavy chain-only antibodies that are one order of magnitude smaller than their full-length counterparts.
Nanobodies have been shown to recognize epitopes that are often inaccessible to conventional antibodies, which could offer advantages in terms of targeting variants.
Another advantage of using nanobodies as antivirals is their convenient bulk production in prokaryotic systems, long shelf-life, and increased tissue permeability.
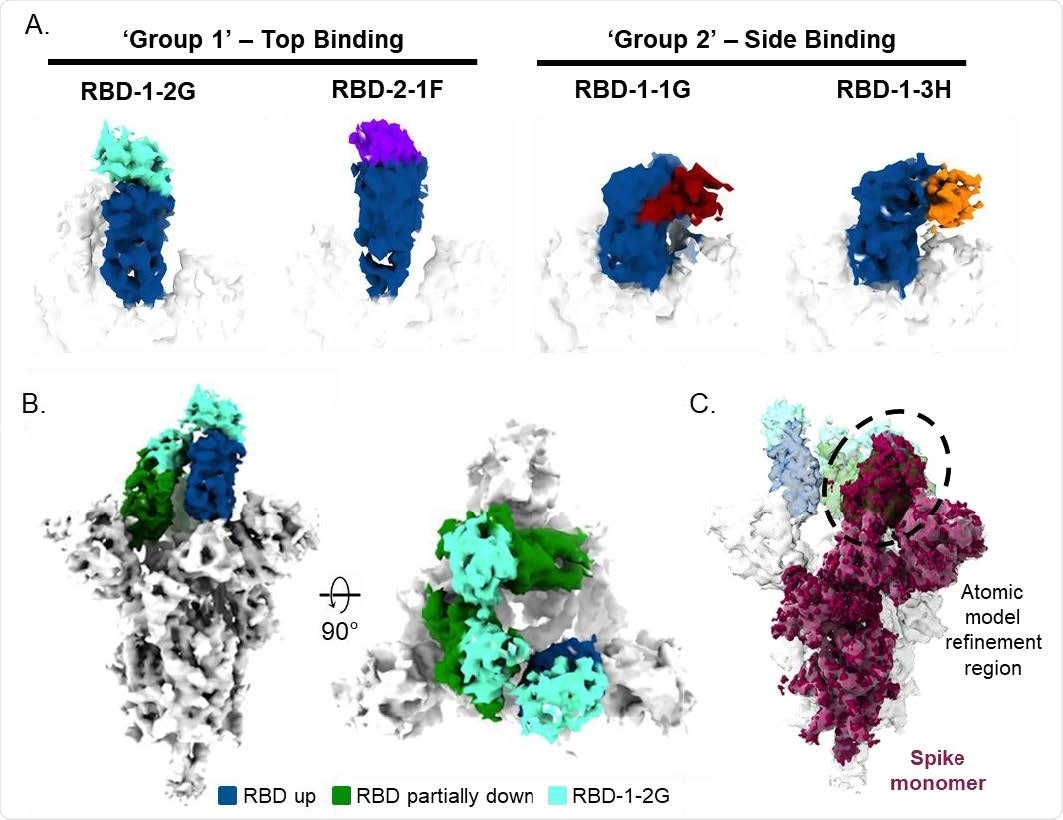
Cryo-EM of SARS-CoV-2 spike trimer and RBD binding nanobodies (A) Cryo-EM analysis of nanobodies complexed with the RBD (up state) revealed two distinct binding modes. (B) Top and side views of the cryo-EM map of S-protein in complex with 3 molecules of RBD-1- 2G (cyan). (C) Side view of the S-protein highlighting the spike monomer region refined during image processing is shown. The density containing the RBD in the laid state was used for the atomic model fitting refinement.
What did the researchers do?
Simeonov and colleagues isolated and characterized SARS-CoV-2 neutralizing single-domain antibodies from a pool of humanized phage display libraries.
These nanobodies bound the spike RBD with subnanomolar affinities and neutralized both spike-protein pseudotyped particles and authentic viruses in mammalian cell models of infection.
The most potent nanobody – RBD-1-2G – yielded an improved affinity and neutralization potency when incorporated into bivalent and trivalent modalities.
Cryogenic electron microscopy showed that RBD-1-2G binds an epitope on the RBD that overlaps with the binding site for ACE2.
The RBD-1-2G nanobody was capable of neutralizing pseudotyped particles containing the N501Y mutation, including those containing the alpha variant.
A primary human airway air-liquid interface ex vivo tissue model demonstrated that RBD-1-2G-Fc treatment reduced viral burden following infection with the alpha strain of SARS-CoV-2.
“A tool to mitigate the threat of emerging SARS-CoV-2 variants”
The researchers say the findings support the versatility of RBD-1-2G as a good prototype for designing new candidates that are capable of adapting to different SARS CoV-2 variants.
“This presented strategy will serve as a tool to mitigate the threat of emerging SARS-CoV-2 variants,” they write.
Simeonov and colleagues say the ability to rapidly screen artificial humanized nanobody libraries to identify compatible binders helps to counter the immunogenic defenses developed by viral pathogens.
“The method described in this study could also be expanded to infectious diseases other than COVID-19 and serve as a technical reserve for rapid neutralizing nanobodies discovery during future pandemics,” concludes the team.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Simeonov A, et al. The humanized nanobody RBD-1-2G tolerates the spike N501Y mutation to neutralize SARS-CoV-2. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.10.22.465476,
- Peer reviewed and published scientific report.
Fu, Ying, Juliana da Fonseca Rezende e Mello, Bryan D. Fleming, Alex Renn, Catherine Z. Chen, Xin Hu, Miao Xu, et al. 2022. “A Humanized Nanobody Phage Display Library Yields Potent Binders of SARS CoV-2 Spike.” Edited by Juan Pablo Jaworski. PLOS ONE 17 (8): e0272364. https://doi.org/10.1371/journal.pone.0272364. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0272364.