The coronavirus disease 2019 (COVID-19) pandemic caused by SARS-CoV-2 has led to hundreds of millions of infections worldwide and almost 5 million deaths. SARS-CoV-2 undergoes frequent mutations of its genome, resulting in the emergence of numerous variants. Some of these are more infectious than the wild-type variant and are called VOCs.
Four notable SARS-CoV-2 VOCs include the Alpha, Beta, Gamma, and Delta variants. Each of these strains of SARS-CoV-2 is more transmissible and/or more virulent than the wild-type and also possesses immune evasion capabilities. The current study examines the virus-receptor interactions at the molecular level.
The researchers modeled the complex formed by the SARS-CoV-2 receptor-binding domain (RBD) and the ACE-2 binding site using the crystal structure. Each VOC exhibits mutations at specific RBD sites on the spike protein. Using four stable RBD-ACE-2 structures, the researchers incorporated substitutions at specific amino acids to give rise to alternative conformations of the RBD.
These structures agree well with those obtained by cryo-electron microscopy, with the mutations occurring near the binding interface of the RBD-ACE-2 complex.
Study findings
The researchers modeled the wild-type, Alpha, Beta, Gamma, and Delta strains of SARS-CoV-2 to assess the root mean square deviation (RMSD). While the RMSD was similar for the Beta and Gamma VOCs, being markedly higher than that of the wild-type virus, the Alpha and Delta are more similar to the wild-type in their RMSD values.
The Root Mean Square Fluctuation (RMSF) of the ACE-2 receptor was then analyzed at stability. This molecule, which is both a viral receptor and a cardiometabolic regulator forms an alpha-helix that interacts at the N-terminal domain (NTD) with the spike RBD. The various key residues such as Glu329, Asn330, and Lys353 form hydrogen bonds or salt bridges with the viral RBD.
Here, differences were found at the NTD and other residues for the Beta and Gamma VOCs. These also showed somewhat higher peak dynamics for the RBD. The spike RBD contains a receptor-binding motif (RBM), where the ACE-2-RBD contact actually occurs, comprising residues 438-506. The RBM residues lie closer to the C-terminal domain (CTD) of the RBD, without major fluctuations per variant.
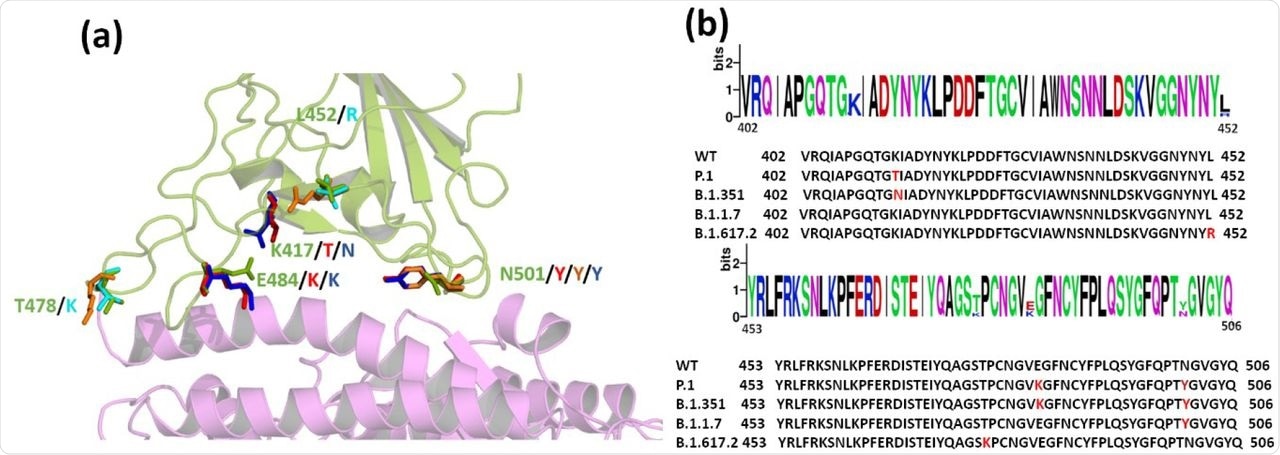 (a) Superimposed structure of the WT and the Variants highlighting the mutations in RBD. Wild type ACE2 receptor is shown in magenta and RBD in green cartoons. The mutations are shown as sticks. (WT: green, P.1: red; B.1.1.7: Orange; B.1.351: blue; B.1.617.2: Cyan) (b) Sequence alignment of WT and the VOCs. Mutated residues are highlighted in red in the sequence
(a) Superimposed structure of the WT and the Variants highlighting the mutations in RBD. Wild type ACE2 receptor is shown in magenta and RBD in green cartoons. The mutations are shown as sticks. (WT: green, P.1: red; B.1.1.7: Orange; B.1.351: blue; B.1.617.2: Cyan) (b) Sequence alignment of WT and the VOCs. Mutated residues are highlighted in red in the sequence
When superimposed on the ACE-2 receptor, the differences in the relative positions of the RBD and ACE-2 were observed by variant. While the Alpha variant shows little difference, the Beta and Delta appear to have moved closer. Comparatively, the Gamma RBD is farther away from the ACE-2 when compared to that of the wild-type strain.
Moreover, both the Beta and Delta VOCs show very different RBD orientations within the RBD-ACE-2 complexes, while the Alpha and Gamma RBDs have similar orientations. This type of shift ultimately affects protein-protein interactions (PPIs) and protein dynamics at the interface.
Indeed, the dominant motion of the RBD with respect to the receptor was seen to be similar for the wild-type, Beta, and Delta VOCs; however, the Gamma and Alpha variants form a separate cluster. To resolve the incongruence with the earlier structural studies where the Alpha, Gamma, and wild-type strains had similar orientations, the researchers studied the energetic characteristics of each complex.
The wild-type RBD was found in two different forms, thus representing its different conformations after binding to the receptor. With the Alpha and Delta variants, the ACE-2 conformations in the complexes remained low-energy, with a greater number of conformations with the Beta and Gamma variant complexes. The scientists attribute this to the fact that the mutations defining various VOCs affect mostly the RBD, leaving large parts of the spike intact, thus preserving the ACE-2 dynamics more or less.
The RBDs of all complexes belonged to one of two complexes. The Alpha and Gamma variants, particularly the former, were found to be very different from each other in protein dynamics as they formed two clusters and one cluster at low energy, respectively. The Beta and Delta variants had shown very different structures from the wild-type on superimposition; however, their dynamics were very similar with each other and the wild-type for both RBD and ACE-2 components.
The Gamma VOC showed the greatest flexibility for both components of the complex, and the Delta variant the least, indicating that it is the most tightly bound of the five.
The binding energies are highest with the Delta and least with the Alpha variant. Notably, both the Delta and Beta variants are the most compact of the systems. The Alpha variant also shows increased accessibility of the RBD to solvent, indicating that the complex undergoes a conformational change.
The residues at the interface govern the engagement and stability of the complexes formed. With the Gamma variant, only Glu484 plays a significant role in binding. Comparatively, the E484K mutation is found in both this and the Beta variant, thus resulting in higher binding energy. The corresponding residue in the wild-type and Alpha variant interacts unfavorably, as shown by the high repulsive energy values.
The L452R and T478K mutations in the Delta variant also increase interaction energies markedly, like the E484K. These changes result in lower binding energies at almost all the complementary residues of the ACE-2 receptor in the complex, thereby indicating an impact on the binding efficiency of the receptor.
A mutation at K417 in the Gamma and Beta variants led to a loss of salt bridge formation between the ACE-2 and RBD of these variants as a result of the loss of hydrogen bonding. The Gamma variant had the least hydrogen bonds, along with the Delta and the wild-type having the greatest number. The Delta variant lost two hydrogen bonds at RBD residues Y505 and T500, linking them with ACE-2 residues D37 and E355, respectively.
This is due to the large shift in conformation following RBD-ACE-2 binding in this complex. Thus, the interface residues, particularly the mutated ones, significantly contribute to stabilizing the complex. With the Delta variant, the interaction energy increases, causing the RBD-ACE-2 complex to be compact and tightly bound, largely due to strong PPIs and dynamics that are very similar to those of the wild-type virus.
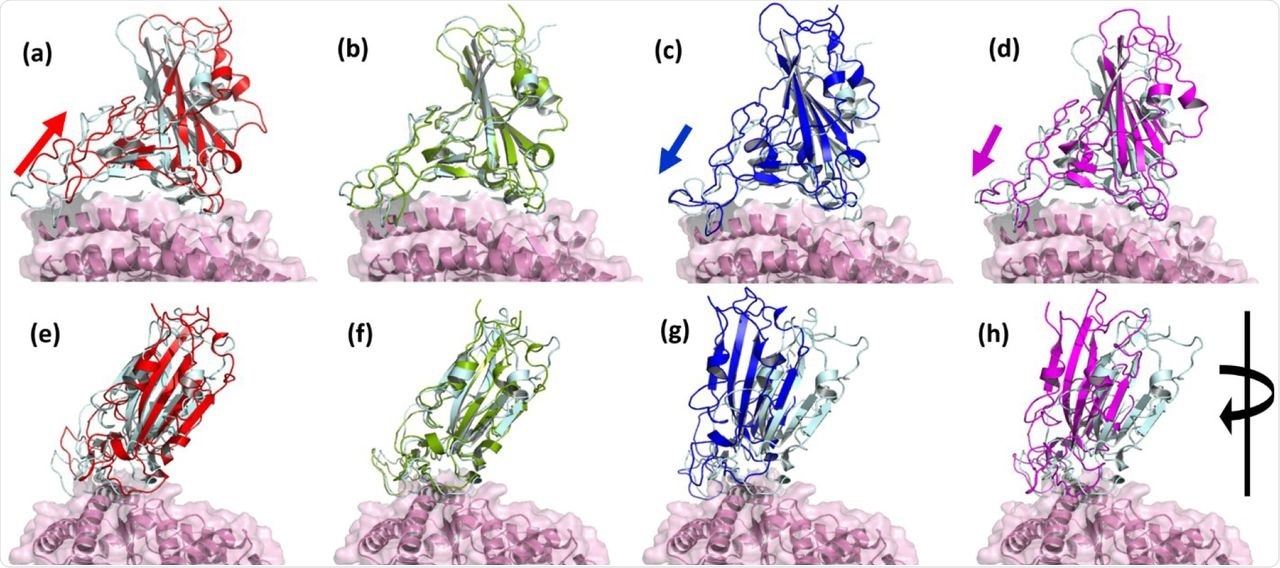 Superimposed structure of WT and VOCs showing the relative variation in the receptor binding domains. Interfacial region of RBD and ACE2 in (a) P.1, (b) B.1.1.7, (c) B.1.351 and (d) B.1.167.2. When the structure is rotated 90° in (e) P.1, (f) B.1.1.7, (g) B.1.351 and (h) B.1.167.2. Relative displacement was observed in B.1.351 and B.1.167.2. (WT: RBD in cyan, ACE2 in pink; P.1 RBD in red; B.1.1.7 RBD in Green; B.1.351 RBD in Blue; B.1.167.2 RBD in Magenta).
Superimposed structure of WT and VOCs showing the relative variation in the receptor binding domains. Interfacial region of RBD and ACE2 in (a) P.1, (b) B.1.1.7, (c) B.1.351 and (d) B.1.167.2. When the structure is rotated 90° in (e) P.1, (f) B.1.1.7, (g) B.1.351 and (h) B.1.167.2. Relative displacement was observed in B.1.351 and B.1.167.2. (WT: RBD in cyan, ACE2 in pink; P.1 RBD in red; B.1.1.7 RBD in Green; B.1.351 RBD in Blue; B.1.167.2 RBD in Magenta).
Implications
The findings of the current study show that the Alpha and Delta variants show little variation in their protein dynamics, despite the presence of several mutations near the RBD-ACE-2 interface. Interacting residues appear to be more closely related to each other with the Beta and Delta variants, though these underwent marked changes in conformation after binding.
The Delta complex is the most tightly bound, with the greatest compactness, and a marked gain in interaction energy. The RBD-ACE-2 binding was reliant on electrostatic interactions to a large extent.
The mutations were the trigger for the greatest changes in interaction energies. With the Beta and Gamma variants, this resulted in both favorable and adverse changes, while there was not much change with the Alpha variant from the wild-type. In the case of the Delta variant, which is currently dominating the global landscape, the interaction energy was drastically increased by the L452R and T478K mutations that are responsible for its compact shape, tight binding, and strong interactions, along with dynamics that are highly similar to the wild-type complex.
When the RBD and ACE-2 have a high affinity for each other, the findings discussed here indicate the gain in viral pathogenicity. These results, therefore, indicate the important role of such studies in designing better therapies to deal with the threat posed by emerging new variants of the virus.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources