Viral infections may have enormous global impacts, as evidenced by the recent coronavirus disease 2019 (COVID-19) pandemic, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). History has many other examples, such as the Ebola epidemics of 2013- 2016, the H1N1 pandemic of 2009-2010, etc. Structural biology aims to determine how antibodies elicited during infection or vaccination target viral proteins. A new review published in the journal Viruses reviews how molecular and structural biology have contributed to our understanding of antibody recognition of HIV-1, SARS-CoV-2, and Zika.
 Study: How Antibodies Recognize Pathogenic Viruses: Structural Correlates of Antibody Neutralization of HIV-1, SARS-CoV-2, and Zika. Image Credit: Fotomay/ Shutterstock
Study: How Antibodies Recognize Pathogenic Viruses: Structural Correlates of Antibody Neutralization of HIV-1, SARS-CoV-2, and Zika. Image Credit: Fotomay/ Shutterstock
A new study
Examples of viruses that have wreaked havoc globally and needed urgent vaccine and therapeutic development are SARS-CoV, Middle East Respiratory Syndrome (MERS), acquired immune deficiency syndrome (AIDS), the Zika virus (ZIKV), and the current SARS-CoV-2. The rapid development of safe and effective vaccines and therapeutics has proven vital to reducing morbidity and mortality from newly emerging viruses.
Structural biologists are continuously advancing techniques in X-ray crystallography and cryo-electron microscopy (cryo-EM) to investigate viruses and viral proteins. In the present study, scientists investigated antibody (Ab) recognition of viruses by solving 3D structures of viral proteins bound to Abs elicited by infection or vaccination. This is essential for the development of effective monoclonal Ab therapies and vaccines.
HIV-1, SARS-CoV-2, and Zika Viruses
Human immunodeficiency virus 1 (HIV-1) is responsible for the AIDS pandemic and has long posed a challenge for vaccine development owing to its striking ability to evade the host immune responses. It contains a single viral protein (Envelope or Env) that facilitates infection. Advances in X-ray crystallography and cryo-EM have made it possible to structurally characterize broadly neutralizing antibodies (bNAb) interactions with Env and Env conformational changes that have subsequently informed vaccine design.
The spike (S) proteins on the surface of SARS-CoV-2 enable it to infect host cells. Structural biology has been instrumental in rapidly characterizing and evaluating the S protein and the antibodies produced in natural infection. This has saved countless lives by contributing to the development of COVID-19 vaccines and monoclonal Ab (mAb) therapeutics.
ZIKV is a mosquito-borne virus that can cause microcephaly and neurodevelopmental abnormalities in the newborns of infected mothers. There is not yet a safe and effective vaccine against ZIKV that is universally available.
Main findings
Structural analysis has helped identify neutralizing epitopes on HIV-1, SARS-CoV-2, and ZIKV. Both X-ray crystallography and cryo-EM analyses of viral have provided insights into mechanisms of neutralization by Abs and identified new therapeutic targets. In the case of HIV-1, targets of bNAbs can be divided into multiple categories, each presenting a distinct landscape for bNAb binding and posing different challenges for Abs to overcome. The mode of binding for bNAbs at all epitopes has been highlighted in structural biology.
X-ray crystallographic and cryo-EM structures of Ab:Env complexes are key to characterizing the binding mode of bNAbs and understanding the context of atypical features. These data have aided in the structure-based design of gp120 and SOSIP-based immunogens, which elicit responses to particular epitopes and design small molecule drugs.
In the case of SARS-CoV-2, the S protein comprises three identical subunits, each containing an RBD. Antibodies that recognize the RBD are a vital part of the neutralizing Ab response. While scientists have mainly focused on the RBD, a growing number of neutralizing Abs that target other regions of the S protein are being found (e.g., NTD and S2 domain). Additionally, some of these Abs are also broadly cross-reactive to other betacoronaviruses.
The E protein of ZIKV is key for facilitating cellular entry and fusion, making it an important target of nAbs that effectively clear ZIKV. Structural characterization of Abs that bind the ZIKV E protein has revealed multiple epitopes. Some Ab epitopes (characterized by crystallography) are not accessible on the known cryo-EM structures of mature ZIKV as evidence suggests that E proteins are dynamic and sample different conformations. It must be noted that the phenomenon of flavivirus “breathing” may be an outcome of the conformational changes of the E protein during the viral life cycle. However, the breathing conformation of ZIKV has not yet been determined.
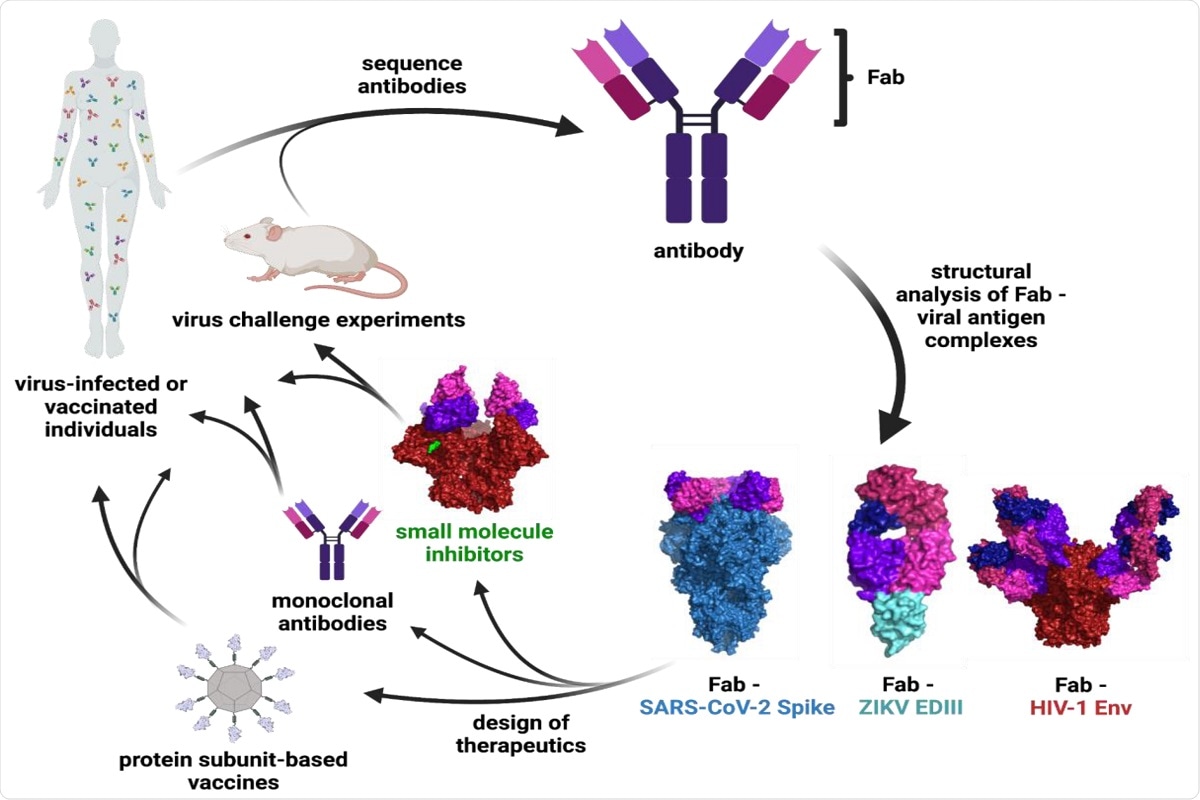 Figure 1: Schematic of Ab characterization and therapeutic development. The binding epitopes of Abs isolated from infected or vaccinated individuals or animal studies are determined through structural analysis of Fab—viral antigen complexes. These structures inform the design of vaccines, monoclonal Abs, and small molecule therapeutics that can be tested in clinical trials and animal models. Surface representations are shown for the following structures: Fab—SARS-CoV-2 S (PDB 7K90), Fab—ZIKV EDIII (PDB 5VIG), Fab—HIV-1 Env (PDB 5T3Z), and small molecule inhibitor—HIV-1 Env (PDB 7LO6).
Figure 1: Schematic of Ab characterization and therapeutic development. The binding epitopes of Abs isolated from infected or vaccinated individuals or animal studies are determined through structural analysis of Fab—viral antigen complexes. These structures inform the design of vaccines, monoclonal Abs, and small molecule therapeutics that can be tested in clinical trials and animal models. Surface representations are shown for the following structures: Fab—SARS-CoV-2 S (PDB 7K90), Fab—ZIKV EDIII (PDB 5VIG), Fab—HIV-1 Env (PDB 5T3Z), and small molecule inhibitor—HIV-1 Env (PDB 7LO6).
Conclusion
Structural biology has allowed for a deeper understanding of the immune responses to viruses, such as HIV-1, SARS-CoV-2, and ZIKV. Structures of Abs bound to viral proteins have increased our understanding of the role of features that Abs develop in response to antigens. Using structural and molecular biology methods, scientists have made tremendous progress towards Ab treatments and vaccines for the HIV-1, SARS-CoV-2, and ZIKV viruses.
Journal reference:
- Abernathy, M. et al. (2021) "How Antibodies Recognize Pathogenic Viruses: Structural Correlates of Antibody Neutralization of HIV-1, SARS-CoV-2, and Zika", Viruses, 13(10), p. 2106. doi: https://doi.org/10.3390/v13102106