The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which causes coronavirus disease 2019 (COVID-19), is a ribonucleic acid (RNA) virus that is prone to genetic mutations. The mutations that accumulate in the virus over time may result in the appearance of multiple variants whose characteristics may vary when compared to the original strain. Adaptive mutations that result from a single amino acid change can cause drastic changes in the pathogenicity of the virus.
Moreover, the emergence of SARS-CoV-2 variants creates a constant need to improve existing vaccine therapies. The B.1.351 variant of SARS-CoV-2, which is also referred to as the Beta variant, caused the second wave of COVID-19 cases in South Africa in October 2020.
 Study: Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants. Image Credit: Design_Cells / Shutterstock.com
Study: Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants. Image Credit: Design_Cells / Shutterstock.com
It has been reported that the SARS-CoV-2 Beta variant exhibits increased transmissibility, as well as decreased neutralization by currently available monoclonal antibodies, serum from convalescent individuals, and post-vaccination sera. In a recent Nature Communications study, the researchers identify and characterize monoclonal antibodies (mAbs) against SARS-CoV-2 receptor-binding domain (RBD) that are effective against the wild-type SARS-CoV-2 strain and its B.1.351 variant.
The neutralizing activity of mAbs 58G6, 510A5, and 13G9
The scientists in the present study identified 20 neutralizing antibodies exhibiting high affinity for the SARS-CoV-2 receptor-binding domain (RBD) by screening blood samples from COVID-19 convalescent individuals using a rapid neutralizing Abs screening system. The neutralizing potencies of the mAbs were determined using their half inhibition concentrations (IC50) against the wild-type SARS-CoV-2 strain.
Further, the scientists selected ten best-performing antibodies from the twenty they identified and evaluated their neutralizing potency by employing the plaque-reduction neutralization testing (PRNT) assay against the wild-type SARS-CoV-2 strain and B.1.351 variant. The antibodies 58G6, 510A5, and 13G9 exhibited good neutralizing efficacy against SARS-CoV-2 with IC50 values ranging from 1.285 to 9.174 ng/mL.
Antibodies 58G6 and 510A5 also showed good neutralizing efficacy against the B.1.351 without being affected by the RBD escape mutation. Most of the ten best performing mAbs identified in this study exhibited neutralizing activity against the B.1.351 variant.
Notably, 58G6 exhibited a similar binding affinity to the S1 subunit of the B.1.351 and SARS-CoV-2 wild-type strains. The antibodies 510A5 and 13G9 showed higher binding affinity to the S1 subunit of the wild-type strain of SARS-CoV-2 when compared to B.1.351 S1 subunit. Most of the 20 neutralizing antibodies identified did not show cross-reactivity to the spike proteins of SARS-CoV or the Middle East respiratory syndrome coronavirus (MERS-CoV).
The study successfully demonstrated that the three mAbs 58G6, 510A5, and 13G9 exhibited high neutralizing efficacies against the original SARS-CoV-2 strain and the B.1.351 variant.
13G9e and 510A5e as epitopes for potent neutralizing mAbs
The antigenic sites on SARS-CoV-2 were assessed to understand the high neutralizing potential of the identified mAbs. Competitive enzyme-linked immunosorbent assay (ELISA) was performed with the best performing 20 mAbs and 54 other mAbs.
The mAbs were segregated into five groups based on their recognition sites on the RBD. It was found that the epitope region for most of the potent neutralizing antibodies overlapped with the epitope for 13G9 (13G9e).
Interestingly, competitive ELISA and surface plasmon resonance (SPR) assay using the best performing 20 mAbs showed that they directly disrupt the interaction between the RBD region of SARS-CoV-2 and the angiotensin-converting enzyme 2 (ACE2) receptor on the host cell.
Competitive ELISA was performed by employing biotinylated mAbs to identify whether the epitopes are interrelated. The scientists found that 16 of the top 20 mAbs competed with 13G9, thus indicating that antigenic sites overlapped with 13G9e. However, the antigenic sites of 510A5, 55A8, 57F7, and 07C1 overlapped with 510A5e.
These findings suggest two independent classes of epitopes on the SARS-CoV-2 RBD, of which 13G9e is a vital epitope for potent neutralizing mAbs against RBD.
Neutralizing antibody cocktail as an effective COVID-19 therapy
The scientists investigated the synergistic effect of the mAbs identified against SARS-CoV-2. They combined one of the three potent neutralizing antibodies 58G6, 510A5, or 13G9 with one of the mAbs that have very low neutralizing potential.
Interestingly all tested antibody combinations showed a synergistic effect against the authentic SARS-CoV-2, thereby indicating the benefit of employing neutralizing Ab cocktails as therapy against COVID-19.
The potent mAb 58G6 retains its affinity to the RBD of B.1.351 variant
The regions on the RBD at which the potent neutralizing Abs interact were identified by performing peptide ELISA with synthesized peptides. It was found that one of the most potent neutralizing mAb, 58G6, recognized the linear region containing amino acids 450–457 of SARS-CoV-2 S (S450–457 ) in the RBD.
A unique hydrogen bond was identified between RBD and 58G6 in this region that played a role in the selective recognition of this linear region by 58G6 and not by 13G9. Both 13G9 and 58G6 recognized the epitope S470-495 that has been reported as an important region recognized by ACE2 and several potent neutralizing anti-SARS-CoV-2 antibodies.
Cryo-electron microscopy (cryo-EM) analysis was performed, which identified a hydrogen bond interaction between N94 and carbonyl group on the main chain of SE484 instead of its side chain. Mutations such as SE484K found in this region in the case of variants like B.1.351 or P.1 will not disrupt the affinity of 58G6 to the RBD.
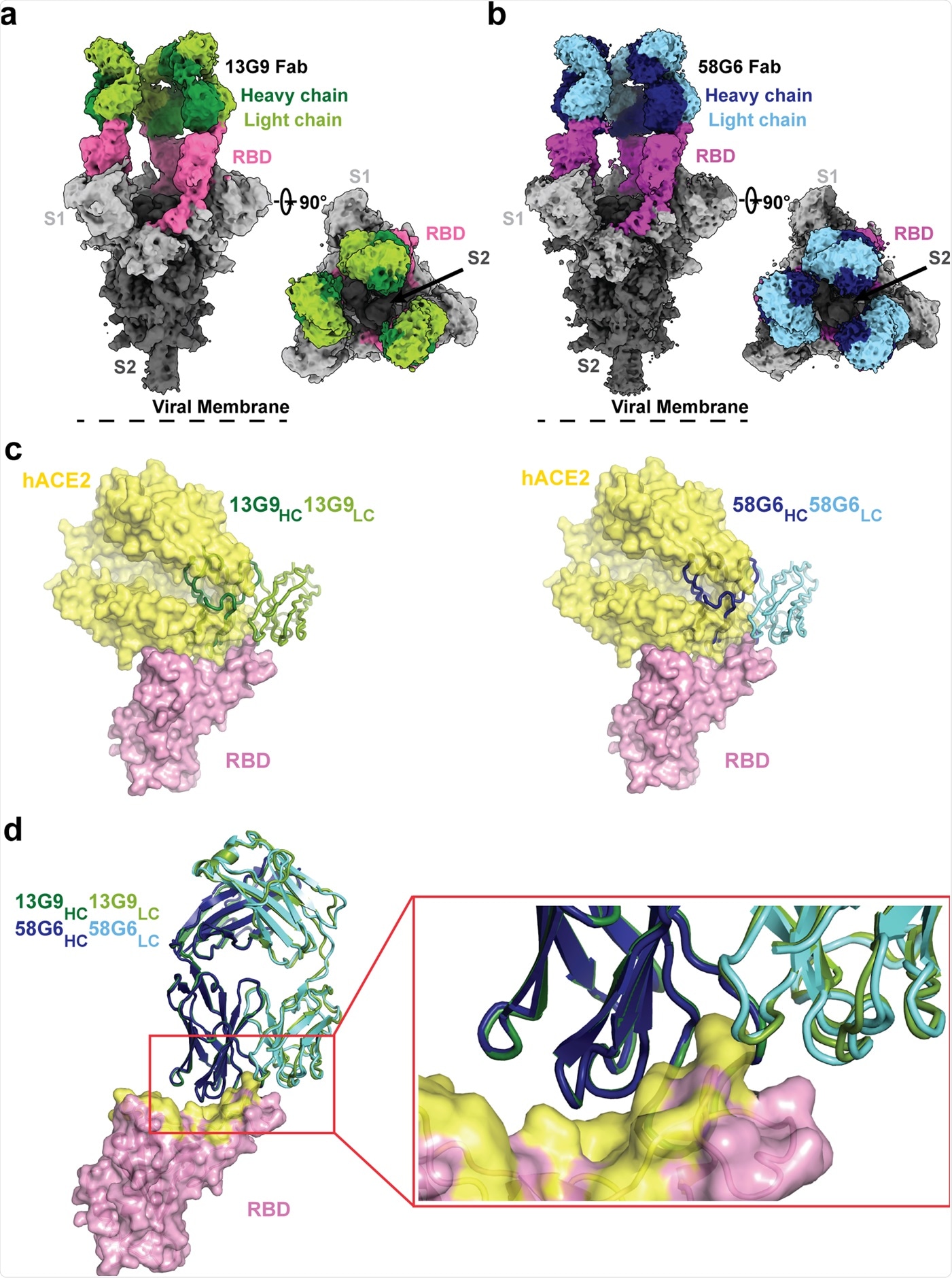 Cryo-EM densities for 13G9 Fab (antigen binding fragment)-S (a; 3.9 Å) and 58G6 Fab-S (b; 3.6 Å) complexes, revealing binding of 13G9 or 58G6 to RBDs in the all ‘up’ state. c Superimposition of RBD-hACE2 [Protein Data Bank (PDB) ID 6LZG][https://www.rcsb.org/structure/6LZG] complex structure together with RBD-13G9 Fab (left) or RBD-58G6 Fab (right) variable domains, respectively. d Alignment of 13G9 and 58G6 Fabs on the same RBD. HC, heavy chain; LC, light chain. CDR, complementarity determining region.
Cryo-EM densities for 13G9 Fab (antigen binding fragment)-S (a; 3.9 Å) and 58G6 Fab-S (b; 3.6 Å) complexes, revealing binding of 13G9 or 58G6 to RBDs in the all ‘up’ state. c Superimposition of RBD-hACE2 [Protein Data Bank (PDB) ID 6LZG][https://www.rcsb.org/structure/6LZG] complex structure together with RBD-13G9 Fab (left) or RBD-58G6 Fab (right) variable domains, respectively. d Alignment of 13G9 and 58G6 Fabs on the same RBD. HC, heavy chain; LC, light chain. CDR, complementarity determining region.
In SPR assays, the 58G6 mAb was shown to retain its affinity to the S1 subunit of the B.1.351 spike protein. However, the mAb 13G9 was unable to retain its affinity to bind to the RBD of the variants.
The SE484K mutation in the RBD of the variant confers a positive charge around R64 present in the complementarity determining region L3 (CDRL3), thus resulting in electrostatic repulsion and decreased affinity for 13G9 binding to the B.1.351 RBD. A slight reduction in the neutralizing effect was observed for 13G9 against the B.1.351 strain.
Notably, very little sequence similarity was observed between the epitope recognized by 13G9 or 58G6 on the SARS-CoV-2 RBD and the corresponding region in SARS-CoV. It was observed that the region was absent in MERS-CoV. This explains the absence of 13G9 or 58G6 cross-reactivity towards SARS-CoV and MERS-CoV.
Further, the study also found that in the case of the potent mAbs 13G9 or 58G6, the RBDs were in the ‘up’ state while interacting with the fragment antigen-binding region (Fabs) of the mAbs, thus resulting in inaccessibility of the RBDs by ACE2. Notably, the mAbs 13G9 and 58G6 were isolated from blood samples of different donors.
However, it was observed that both of these mAbs were transcribed from the variant regions IGHV1-58 and IGKV3-20. This indicates the importance of the pairing of germline genes of IGHV1-58 and IGKV3-20 for neutralizing SARS-CoV-2 and associated variants.
58G6 and 510A5 exhibited positive outcomes in the hACE2 transgenic mouse model
The mAbs 58G6 and 510A5 exhibited potent neutralizing potential against B.1.351. As a result, their efficacy was further investigated in hACE2-transgenic mice.
For this experiment, the mice were administered the antibodies 58G6 and 510A5 or phosphate-buffered saline (PBS) as intraperitoneal injections and subsequently exposed intranasally to the wild-type strain fo SARS-CoV-2 or the B.1.351 variant. It was found that both 58G6 and 510A5 treated groups of animals exposed to the wild-type SARS-CoV-2 strain were able to maintain their body weight for three days post-infection.
Similarly, in the case of animals exposed to B.1.351, both 58G6- and 510A5-treated groups showed a small amount of weight loss on days one and two after infection. However, by day three, the weight loss was successfully curtailed by the mAbs. Further, SARS-CoV-2 and B.1.351 viral load were observed to be reduced after a single dose of 58G6 or 510A5.
These findings suggest that mAbs 58G6, and 510A5 were effective against both wild-type SARS-CoV-2 and B.1.351 infection in hACE2 transgenic mice, thus indicating their potential as a prophylactic agent for COVID-19.
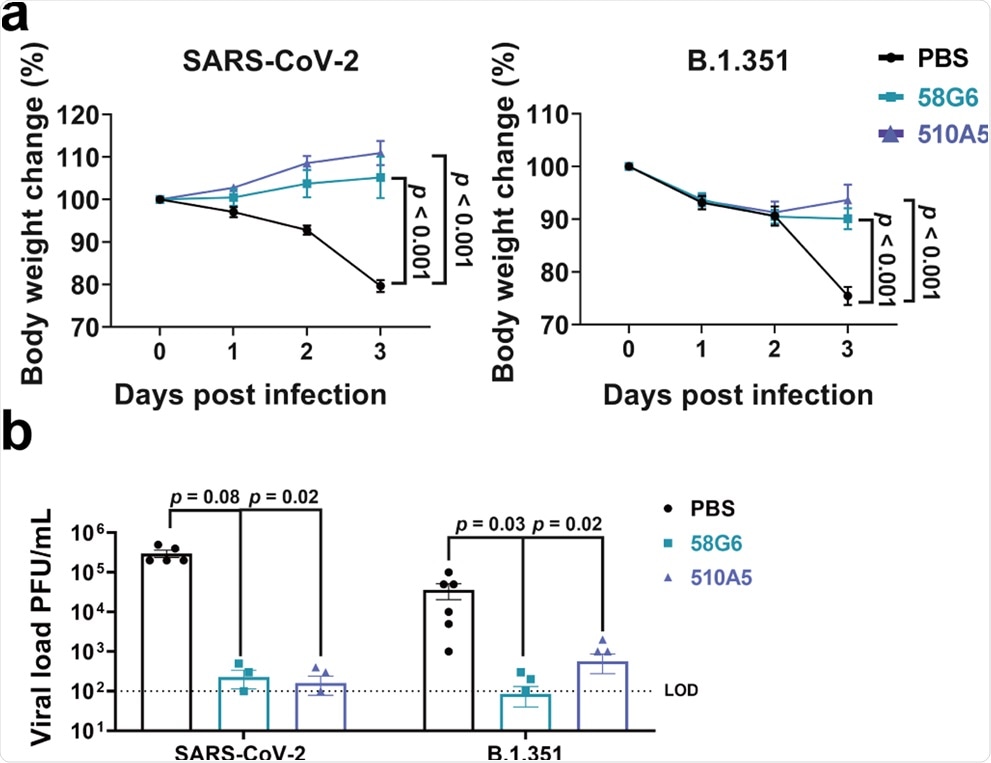 Body weight changes were recorded for PBS (SARS-CoV-2 (WIV04): n = 5; B.1.351: n = 6), 58G6 (SARS-CoV-2 (WIV04): n = 4; B.1.351: n = 7) and 510A5 (SARS-CoV-2 (WIV04): n = 5; B.1.351: n = 7) treatment groups. All the mice received one dose of antibodies (10 mg/kg body weight) injected (i.p.) 24 h prior to the intranasal challenge with SARS-CoV-2 (WIV04) (left) or B.1.351 (right). Equal volume of PBS was used as negative control. The weight loss was recorded over a course of 3 days. b The virus loads in infected lungs of the PBS group, the 58G6 group and the 13G9 group as shown in a were determined by PRNT at 3 days post infection (dpi). The mean values ± SEM of all data points were shown. p values were calculated via two-sided Student’s t-test. Dashed line indicated assay limits of detection (LOD).
Body weight changes were recorded for PBS (SARS-CoV-2 (WIV04): n = 5; B.1.351: n = 6), 58G6 (SARS-CoV-2 (WIV04): n = 4; B.1.351: n = 7) and 510A5 (SARS-CoV-2 (WIV04): n = 5; B.1.351: n = 7) treatment groups. All the mice received one dose of antibodies (10 mg/kg body weight) injected (i.p.) 24 h prior to the intranasal challenge with SARS-CoV-2 (WIV04) (left) or B.1.351 (right). Equal volume of PBS was used as negative control. The weight loss was recorded over a course of 3 days. b The virus loads in infected lungs of the PBS group, the 58G6 group and the 13G9 group as shown in a were determined by PRNT at 3 days post infection (dpi). The mean values ± SEM of all data points were shown. p values were calculated via two-sided Student’s t-test. Dashed line indicated assay limits of detection (LOD).
Conclusion
The present study has identified two potent mAbs against the SARS-CoV-2 RBD, which demonstrates significant efficacy against the B.1.351 variant. Notably, the majority of the reported neutralizing Abs are less effective against this variant.
The two identified antibodies also exhibited good prophylactic efficacy in hACE2-transgenic mice. The findings from this study suggest that the two broad-spectrum mAbs against SARS-CoV-2 RBD hold the potential to evolve as effective prophylactic therapy against SARS-CoV-2 variants.
Journal reference:
- Li, T., Han, X., Gu, C., et al. (2021). Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants. Nature Communications. doi:10.1038/s41467-021-26539-7.