Scientists from Northwestern University, USA, have recently developed a high-throughput, automated screening platform to rapidly identify anti-severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antibodies. The study is currently available on the bioRxiv* preprint server.
Screening platforms currently used for antigen-specific antibody identification utilize directed evolution or isolation of single B-cell clones from COVID-19 recovered individuals or infected animals. The isolation, evaluation, and identification of the best antibody candidate require a series of time-consuming and labor-intensive experiments, including cloning, transfection, cell-based protein expression, protein purification, and binding assessment. The turnaround time of these procedures is weeks to months. Moreover, these procedures often exhibit low efficacy in identifying potent neutralizing antibodies against SARS-CoV-2.
In the current study, the scientists have developed an automated antibody discovery platform that combines cell-free protein synthesis with a high-throughput protein-protein interaction screening.
High-throughput antibody discovery platform
The platform combines four major steps, including cell-free DNA assembly and amplification, cell-free protein synthesis, an amplified luminescent proximity homogeneous linked immunosorbent assay, and an automated workflow utilizing robotic and acoustic liquid handling. The cell-free protein synthesis systems used in the study can generate antibodies directly from linear DNA templates. Similarly, the immunoassay can rapidly characterize protein-protein interaction without requiring protein purification.
The platform takes only 24 hours to screen and characterize hundreds of antigen-specific binding antibodies. For functional validation, the scientists utilized this automated screening platform to test a panel of 120 previously identified antibodies targeting the spike protein of SARS-CoV-2.
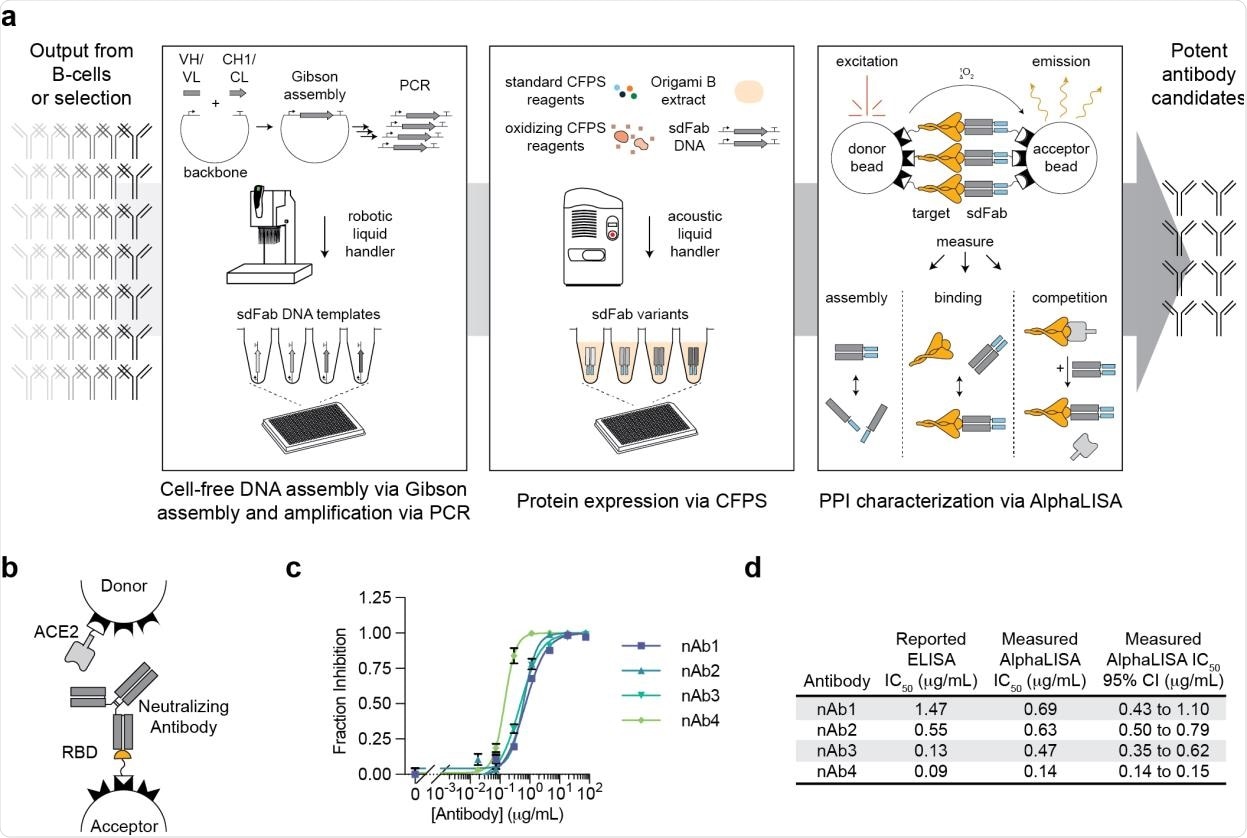
A high-throughput, cell-free antibody screening workflow. a, Schematic of the steps involved in the cell-free antibody screening workflow. b, Diagram of the AlphaLISA screen for neutralizing antibodies via competition with ACE2 for the SARS-CoV-2 RBD. c, Evaluation of commercial neutralizing antibodies (nAbs) in the AlphaLISA ACE2 competition screen (n=3 independent replicates ± SEM). d, Comparison of the reported and measured potencies of commercial neutralizing antibodies.
Detection of protein-protein interaction using high-throughput antibody discovery platform
The binding ability of antibody candidates generated using the cell-free systems was evaluated using the amplified luminescent proximity homogeneous linked immunosorbent assay. This high-throughput screening method can characterize protein-protein interactions directly from cell-free protein synthesis reactions. In addition, the method non-covalently immobilizes proteins of interest on donor and acceptor beads, which produce a chemiluminescent signal upon interactions. Importantly, the technique can characterize direct antibody-antigen binding as well as competitive binding for specific epitopes.
The analysis of five commercially available antibodies revealed that this immunoassay could determine the ability of antibodies to compete with human angiotensin-converting enzyme 2 (ACE2) for binding with spike receptor-binding domain (RBD) of SARS-CoV-2. Further testing with heavy and light chains of the antigen-binding fragment revealed that the assay is highly consistent in predicting antibody assembly.
The efficacy of the immunoassay to characterize antibody binding was further evaluated using distinct panels of antibodies that are known to bind spike trimer or spike RBD or compete with ACE2 for RBD binding. These experiments included a panel of 120 already identified and tested antibodies.
The findings revealed that the amplified luminescent proximity homogeneous linked immunosorbent assay is highly efficient in specifically detecting antibodies that bind to spike trimer, spike RBD, or compete with ACE2 for RBD binding.
Because more than 90% of neutralizing antibodies act by blocking the ACE2 – RBD interaction, the study compared ACE2 competition against virus neutralization. The findings revealed that the assay could consistently identify potent neutralizing antibodies through ACE2 blocking mechanism. However, the assay showed low efficiency in characterizing less-potent neutralizing antibodies.
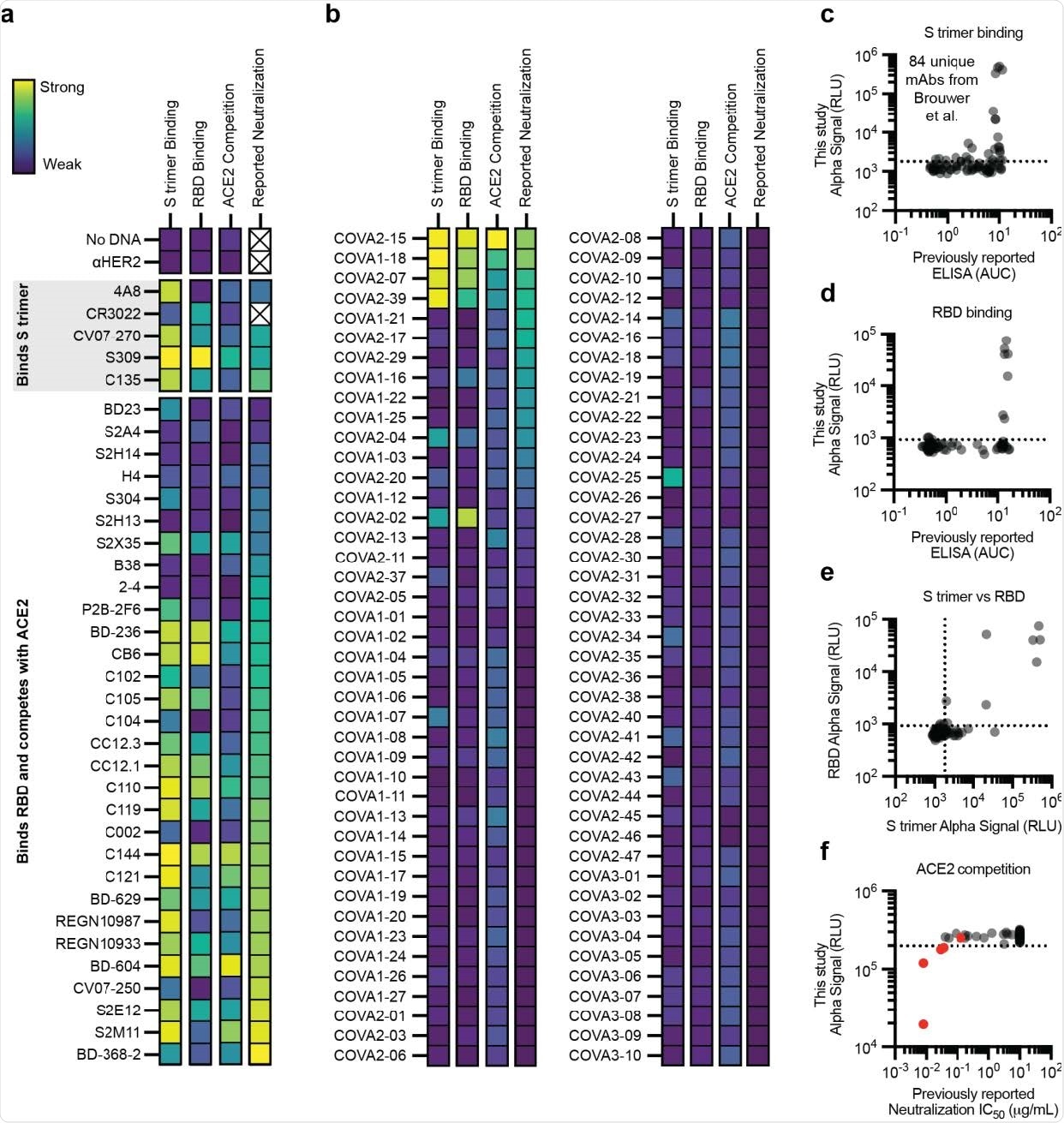
Performance of the cell-free antibody screening workflow evaluated on SARS-CoV-2 neutralizing antibodies. a-f, AlphaLISA data are presented as the mean of 3 independent replicates. A dashed line indicates three standard deviations away from the background signal. a-b, Heatmap of the binding of previously published antibodies measured using AlphaLISA to detect S trimer binding (log10 scaled), RBD binding (log10 scaled), and ACE2 competition (linearly scaled). AlphaLISA data are presented as the mean of 3 independent replicates. The lowest reported neutralization IC50 value is also plotted for comparison (log10 scaled) and an X indicates no relevant data available (Supplementary Table 2). a Heatmap of the binding of 36 diverse antibodies. b, Heatmap of the binding of all 84 antibodies in the Brouwer et al. data set. c-d, Parity plots comparing the AlphaLISA the 84 antibodies in the Brouwer et al. data set vs the published ELISA data. A dashed line indicates three standard deviations away from the background. c, S trimer binding. d, RBD binding. e, Comparison of the S trimer and RBD AlphaLISA binding data. f, Parity plot comparing the AlphaLISA ACE2 competition data for the 84 antibodies in the Brouwer et al. data set vs the published pseudovirus neutralization data. Antibodies that were reported to compete with ACE2 by Brouwer et al. are plotted in red.
Study significance
The study describes the development and validation of a high-throughput, automated antibody discovery platform that utilizes cell-free expression and screening systems. The main advantage of the platform is rapid turnaround time and throughput. For example, a single researcher can characterize a panel of 120 antibodies within 24 hours using this method.
Another significant advantage is the direct profiling of synthesized antibodies using cell-free extracts. This waives the need for protein purification procedures that are often considered the limiting step in antibody screening.
As mentioned by the scientists, this high-throughput platform can be used for rapid and easy identification of potent antibodies for therapeutic, diagnostic, and other basic research applications.