Even though droplets have been seen as the main transmission route for SARS-CoV-2 – a causative agent of the ongoing coronavirus disease (COVID-19) pandemic – it quickly became apparent that airborne transmission also plays a pivotal role in disease spread.
However, the main challenge for understanding airborne transmission has been the inability to reliably examine the structure and dynamics of viruses once they become a part of respiratory aerosol particles.
According to the correct definition, aerosols are smaller than five microns in diameter, and are able to travel significant distances and float in the air for hours (akin to cigarette smoke), and can be inhaled rather easily.
In addition to the different sizes and composition of aerosols, viruses can also be modified to mirror new variants of concern, with various mutations affecting interactions with particular species in the aerosol and subsequently changing its viability and/or structural dynamics.
This is why a recent paper from the Amaro group at the University of California San Diego completely revised contemporary models of viral airborne transmission by providing an unparalleled atomic-level understanding of SARS-CoV-2 within a respiratory aerosol.
Overall schematic depicting the construction and multiscale simulations of Delta SARS-CoV-2 in a respiratory aerosol. (N.B.: The size of divalent cations has been increased for visibility.)

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Developing a ‘computational microscope’
The approach of these researchers relied on the use of all-atom molecular dynamics simulations as a multiscale ‘computational microscope.’ These types of simulations can give rise to multiple types of biological data (such as multiresolution structural datasets) into cohesive and biologically precise structural models.
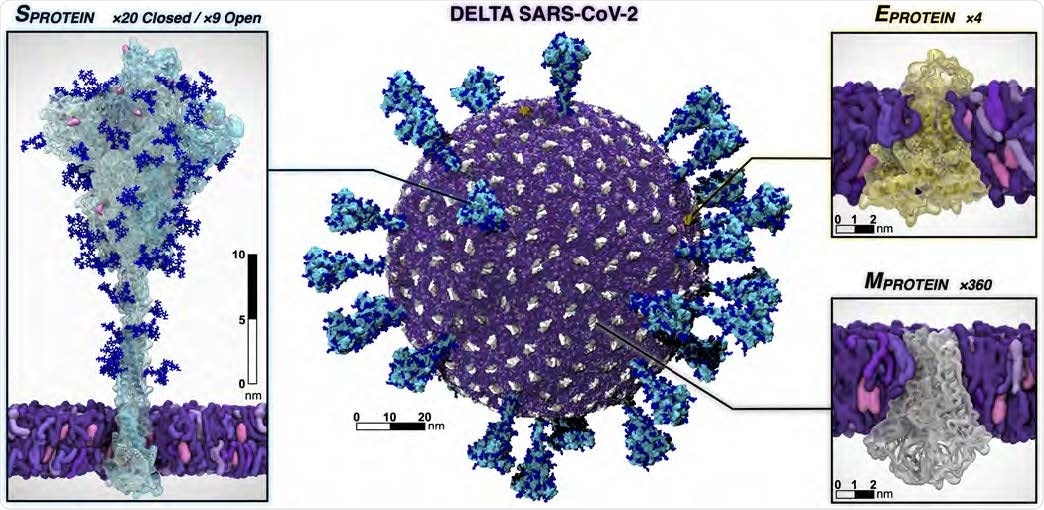
Individual protein components of the SARS-CoV-2 Delta virion. The spike is shown with the surface in cyan and with Delta’s mutated residues and deletion sites highlighted in pink and yellow, respectively. Glycans attached to the spike are shown in blue. The E protein is shown in yellow and the M protein is shown in silver and white. Visualized with VMD.
And once created, the model can then be approximated to its many atoms, developing trajectories of its time-dependent dynamics under aerosol-like conditions. Furthermore, such simulations can also yield specific parameters that can be incorporated into physical models of aerosols.
“Our approach to simulating the entire aerosol follows a composite framework wherein each of the individual molecular pieces is refined and simulated on its own before it is incorporated into the composite model,” study authors further explain their methodological approach.
First atomic-level views of virus-laden aerosols
This study displayed an extensible multiscale computational framework that spans time and length scales from electronic structure via whole aerosol particle dynamics and morphology. In addition, the researchers have also developed all-atom simulations of respiratory mucins to comprehend the structural foundation of interaction with the SARS-CoV-2 spike glycoprotein.
SMA system captured with multiscale modeling from classical MD to AI-enabled quantum mechanics. For all panels: S protein shown in cyan, S glycans in blue, m1/m2 shown in red, ALB in orange, Ca2+ in yellow spheres, viral membrane in purple. A) Interactions between mucins and S facilitated by glycans and Ca2+. B) Snapshot from SMA simulations. C) Example Ca2+ binding site from SMA simulations (1800 sites, each 1000+ atoms) used for AI-enabled quantum mechanical estimates from OrbNet Sky. D) Quantification of contacts between S and mucin from SMA simulations. E) OrbNet Sky energies vs CHARMM36m energies for each sub-selected system, colored by total number of atoms. Performance of OrbNet Sky vs. DFT in subplot B97x-D3/def-TZVP, R2=0.99, for 17 systems of peptides chelating Ca2+ (Hu et al., 2021)). Visualized with VMD.
As a result, this paper puts forth an all-atom simulation that captures massive biological and chemical complexity within a respiratory aerosol. In addition, a described simulation provides the first atomic-level views of aerosols laden with viruses, serving as a basis for developing a plethora of testable hypotheses.
Changes in pH value seen in the aerosol environment may change the dynamics and communication pathways in crucial functional regions of the spike glycoprotein of SARS-CoV-2 (and especially for the Delta strain), while its dramatically open state may facilitate binding to human host cells.
Finally, the study has shown how high-performance computing and cloud resources may speed up scientific efforts and facilitate complex collaborations. Likewise, artificial intelligence can be coupled with high-performance computing at multiple levels to improve effective performance.
Implications for understanding airborne transmission
This work extends the possibility of using multiscale computational microscopy to answer significant questions regarding our understanding of aerosols at the atomic and molecular level, which currently hampers our knowledge of airborne transmission.
“We demonstrate how our integrated data-driven platform provides a new way of exploring the composition, structure, and dynamics of aerosols and aerosolized viruses, while driving simulation method development along several important axes”, say study authors in this bioRxiv paper.
Nonetheless, many technological advances are needed to study even more biologically complex systems that can span from nanometers to one micrometer in size and in longer timescales (i.e., from microseconds to seconds). This is a challenge that needs to be addressed by further studies in the field.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Dommer, A. et al. (2021). #COVIDisAirborne: AI-Enabled Multiscale Computational Microscopy of Delta SARS-CoV-2 in a Respiratory Aerosol. bioRxiv. https://doi.org/10.1101/2021.11.12.468428, https://www.biorxiv.org/content/10.1101/2021.11.12.468428v1
- Peer reviewed and published scientific report.
Dommer, Abigail, Lorenzo Casalino, Fiona Kearns, Mia Rosenfeld, Nicholas Wauer, Surl-Hee Ahn, John Russo, et al. 2022. “#COVIDisAirborne: AI-Enabled Multiscale Computational Microscopy of Delta SARS-CoV-2 in a Respiratory Aerosol.” The International Journal of High Performance Computing Applications, October, 109434202211282. https://doi.org/10.1177/10943420221128233. https://journals.sagepub.com/doi/full/10.1177/10943420221128233.