A recent study conducted by a team of US-based scientists has highlighted a novel function of 3C-like protease of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The protease has been found to cleave a human immune signaling protein called NF-kB Essential Modulator (NEMO) and disrupt the host immune response. The study is currently available on the bioRxiv* preprint server while awaiting peer review.
The non-structural proteins of SARS-CoV-2, including papain-like protease and 3C-like protease, play essential roles in regulating viral lifecycle and disrupting host immune defense. The 3C-like protease recognizes a specific motif in viral polyproteins and cleaves them to form mature non-structural proteins. The same recognition motif is also present in some proteins of the host innate immune pathways. This indicates that 3C-like protease can cleave host proteins to disrupt the first-line antiviral defense mechanism.
In the current study, scientists have investigated whether 3C-like protease has any impact on human immune signaling protein NEMO. During canonical NF-kB response signaling, NEMO cleaves and activates NF-kB, which is an important step for early-stage viral clearance.
NEMO binding to 3C-like protease
The study observed that 3C-like protease interacts with Lys226-His235 of the α-helical Hlx2 domain of one NEMO protomer for cleavage. Thus, as mentioned by the scientists, the protease might utilize a transient state of the α-helical domain for proteolysis. In addition, it might actively outcompete NEMO dimer interactions and unwind the α-helix.
Specifically, 3C-like protease formed hydrophobic interactions and hydrogen bonds with specific residues in the NEMO to outcompete the interactions between these residues in the NEMO dimer.
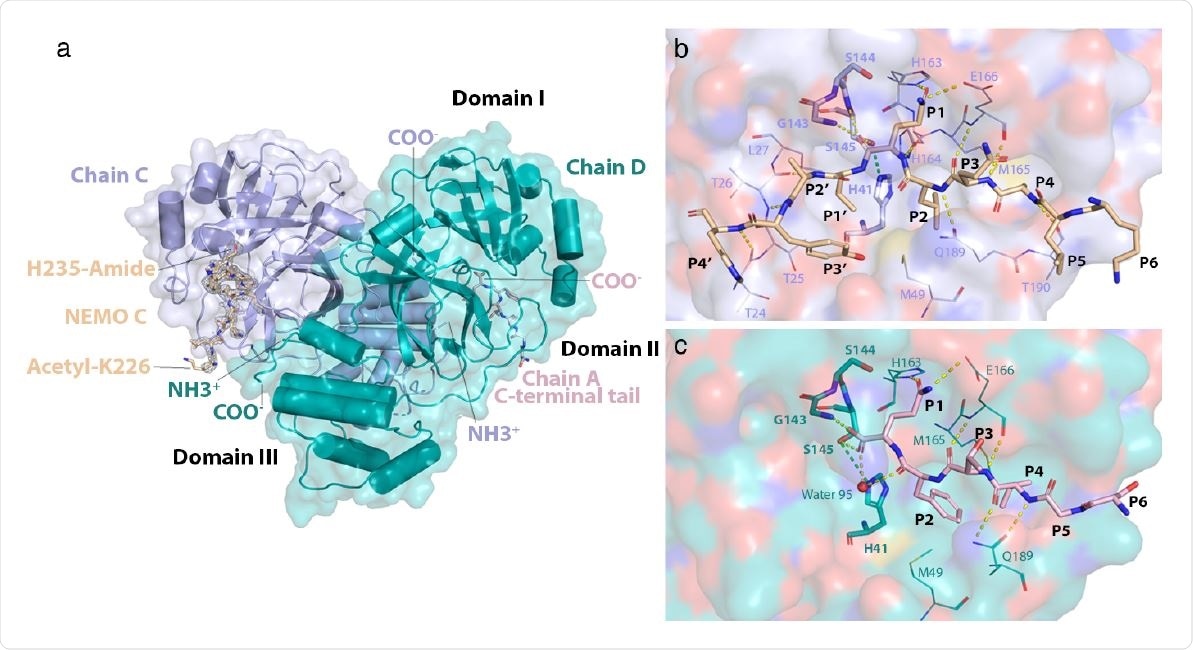
Structure of the NEMO-bound 3CLpro C145S homodimer. a) Global view of NEMObound 3CLpro dimer. Chains C and D have their N- (NH3+) and C-termini (COO-) labeled. NEMO bound into chain C (NEMO C) is displayed as sticks and colored wheat. A mesh of electron density is displayed around NEMO C in light grey. Acetylated Lys226 and amidated His235 at the N- and C-terminus of the NEMO peptide, respectively, are labeled. The C-terminal tail of chain A is colored in light pink, displayed as sticks and is observed to bind into the substrate-binding site of chain D. Domains I (aa. 8-101), II (aa. 102-184) and III (aa. 201-303) are labeled in black. b) Interactions of NEMO with the substrate-binding groove of 3CLpro C145S. NEMO is depicted in wheat. Residues Lys226 to His235 of NEMO are labeled P6 to P4’, respectively. The surface of chain C is shown in light blue and colored by atom type, where oxygens are red, nitrogens are dark blue, carbons are light blue, and sulfurs are yellow. Residues in the substrate-binding site of chain C are portrayed as lines and labeled. Catalytically relevant residues22 in chain C are portrayed as sticks and also labeled in bold. Hydrogen bonds are depicted as dashed yellow lines. The hydrogen bond that would form between Cys145 (in WT) or Ser145 (in C145S) and His41 is depicted as a dashed green line. c) Interactions of the C-terminal tail of chain A with the substrate-binding groove of 3CLpro C145S. The C-terminal tail of chain A is depicted as sticks in light pink. Residues Ser301 to Gln306 are labeled P6 to P1 respectively. The surface of chain D is shown in teal and colored by atom type. Residues in the substrate-binding groove of chain D that interact with the C-terminal tail of chain A are portrayed as lines and labeled. Catalytically relevant residues and hydrogen bonds are portrayed as in (b). The oxygen atom of water 95 is depicted as a red sphere.
Substrate-selectivity of 3C-like protease
The study identified specific subsites that contribute to the substrate versatility of 3C-like protease. Specific interactions of 3C-like protease with its N-terminal autoprocessing sequence or NEMO substrate were identified.
The findings revealed that the S1 subsite is flexible and binds to the NEMO substrate through hydrophobic interactions. However, this subsite did not interact with the N-terminal substrate. In contrast, the S5 subsite interacted with the N-terminal substrate by recruiting a water molecule.
The interactions of S2 subsite with N-terminal and NEMO substrates were found to be conserved. However, the S3 and S4 subsites were identified as significant contributors to substrate versatility.
The findings of the computational analysis revealed that the single substitution S46A in the substrate-binding site of 3C-like protease increases local rigidity, leading to a reduction in NEMO affinity. Similarly, it was predicted that another substitution mutation V232A in NEMO can impair its interaction with 3C-like protease.
Functional consequences of 3C-like protease – NEMO interaction
The enzymatic assay findings revealed that cleavage of NEMO by 3C-like protease is a slow process and does not have a significant functional impact in the initial hours of infection. Moreover, it has been observed in other studies that SARS-CoV-2 proteases cleave multiple host proteins, leading to a significant reduction in protein levels within 24 – 48 of infection.
Overall, these findings indicate that 3C-like protease-mediated cleavage of NEMO is a part of various other mechanisms that are triggered by SARS-CoV-2 to suppress host immune responses.
Given the clinical similarities between NEMO ablation and COVID-19, it could be considered that 3C-like protease-mediated cleavage of NEMO plays a significant role in COVID-19 pathophysiology.
Study significance
The study identifies the role of SARS-CoV-2 3C-like protease in cleaving host immune protein NEMO. It is known that NEMO plays a central role in the mitochondrial antiviral-signaling protein-mediated antiviral immune response. Furthermore, ablation of NEMO is known to cause the inactivation of NF-kB and type 1 interferon response.
Given these observations, the scientists suggest that 3C-like protease can be considered as a potential therapeutic target for preventing virus-induced host immunosuppression as well as viral replication.