To contain the ongoing coronavirus disease 2019 (COVID-19) pandemic, caused by the rapid spread of severe acute respiratory disease coronavirus-2 (SARS-CoV-2), scientists have developed novel vaccines and therapeutics. However, the available COVID-19 vaccines have been designed against the original SARS-CoV-2 strain first reported in Wuhan, China, in late December 2019, and the evolution of the virus has threatened their efficacy. Therefore, it is imperative to develop more effective therapeutics to reduce the rate of infection and mortality caused by new SARS-CoV-2 variants.
The bromodomain and extraterminal domain (BET) family of proteins comprise BRD2, BRD3, BRD4, and BRDT. Characteristic features of the BET protein family members include the presence of two highly conserved N-terminal bromodomains (BDs) and an extraterminal (ET) domain. Previous studies have pointed out that BETs interact with histones and cellular transcriptional machinery.
One of the important roles of BDs is their function as bonafide reader domains of acetylated lysines in histone and non-histone proteins. These proteins have been the molecular targets of small molecule BET inhibitors (e.g., JQ1). Earlier studies have revealed that the ET domain possesses less protein binding properties than BDs.
BET proteins are associated with many cellular functions such as chromatin remodeling, cell proliferation, and gene expression. Among the BETs proteins, BRD4 has been an extensively studied protein, which exists in different splice isoforms, namely, long (BRD4L) and short (BRD4S) isoforms. An intermediate or third isoform had been found only in osteosarcoma cells. Scientists have indicated that BRD2, BRD3, and BRD4 interact with viral proteins of herpesviruses, flaviviruses, and papillomaviruses.
Role of BETs in Viral Infection
Patients infected with SARS-CoV-2 often experience impaired type I interferon (IFN-I) responses and overproduction of proinflammatory cytokines. BRD4 has been recognized as an antiviral gene and also a potent co-activator of proinflammatory cytokines. In this context, scientists have reported that BRD4 coactivates interferon-stimulated genes (ISGs) in the lungs during viral infection by employing P-TEFb. Additionally, it also triggers pro-inflammatory responses in chronic obstructive pulmonary disease, lung fibrosis, and asthma.
Interestingly, a prior study has shown that BET inhibitors can successfully attenuate transcriptional activation of the antiviral response in influenza A infection. In addition, a recent study has identified BRD4 and BRD2 as high confidence interactors of the SARS-CoV-2 E protein.
Most importantly, BRD2 functions as a transcriptional regulator of angiotensin-converting enzyme 2 (ACE2), which is the entry point of the SARS-CoV-2 virus. These studies have also shown that knocking out the BRD2 gene or prophylactic application of BET inhibitors decreases ACE2 expression and, hence, reduces viral infection.
Considering the past shreds of evidence on BRD2 and BRD4 associated with viral infections, researchers have recently evaluated the function of all relevant BET proteins during COVID-19 infection. In this context, they have reported that inhibition of BET proteins post virus invasion or knockout of BRD3 or BRD4 genes in cells leads to overexpression of ACE2, which significantly enhances viral replication. This study is available on the bioRxiv* preprint server.
SARS-CoV-2 E Protein and BET Protein of the Host
In this study, researchers have suggested that BET proteins (BRD4 > BRD3 > BRD2) have prominent antiviral functions, particularly after the SARS-CoV-2 invades the host cell. Furthermore, BET proteins could inhibit COVID-19 infection at the post-entry stage because these proteins are essential for full induction of the type I interferon response and the IL-6 proinflammatory cytokine. In addition, researchers stated that viral replication intensifies after the inactivation of BET proteins via a chemical or genetic method.
The present study revealed a new function of the SARS-CoV-2 E protein, i.e., it antagonizes interferon and ISG expression. Notably, researchers reported that viral E protein in an acetylated form can effectively prevent the antiviral function by binding to the second bromodomain of BRD4. This study reported that among the BET proteins, BRD2 is the most pro-viral owing to its ability to positively regulate the expression of ACE2.
Additionally, BRD4 has a robust antiviral function due to its co-activator role in the induction of interferon genes. Although BRD2 and BRD4 share 70% sequence similarity in their N-terminus, the differences lie in their domain structure.
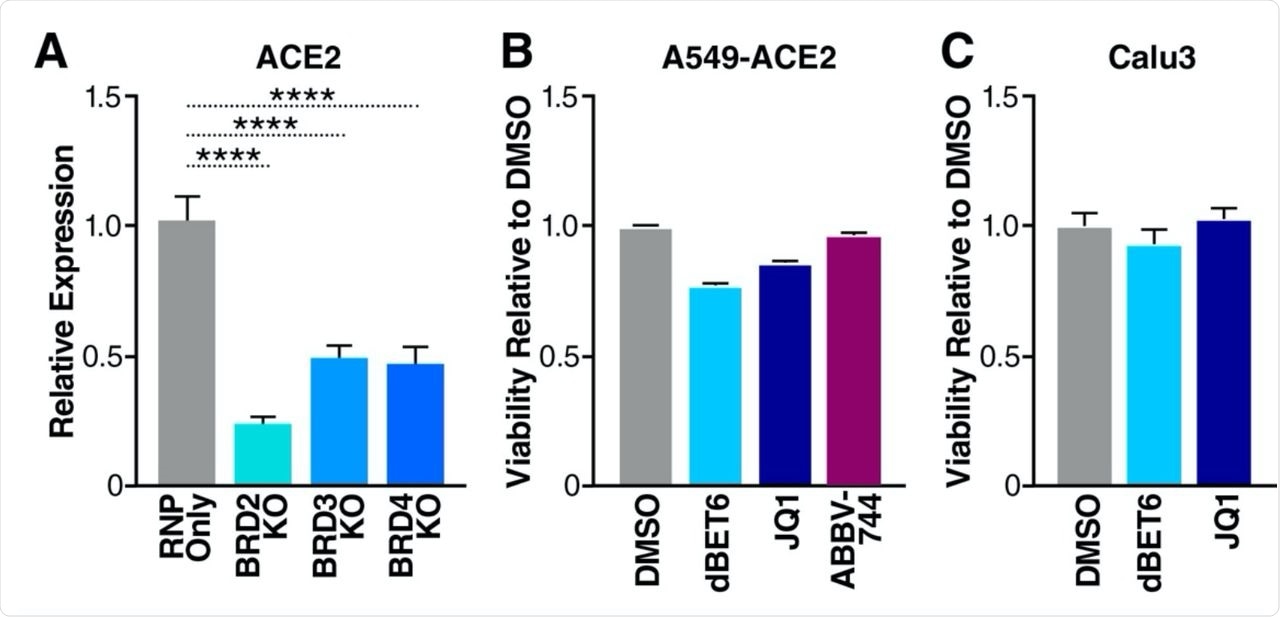
BET proteins are positive regulators of ACE2 expression A. RT-qPCR of ACE2 RNA isolated from Calu3 cells with indicated BET KOs. Data are expressed relative to RNP Only cells. Average of three independent experiments analyzed in triplicate ± SEM are shown and compared to RNP Only samples by ANOVA: ****p<0.0001. B. Viability of A549-ACE2 cells treated with DMSO (vehicle), JQ1 (500nM), dBET6 (500nM), and ABBV-744 (500nM) for 48 hours relative to DMSO. C. Viability of Calu3 cells treated with DMSO (vehicle), JQ1 (500nM), and dBET6 (500nM) for 48 hours relative to DMSO.
Scientists have also found increased viral replication, hypothermic and hyperthermic conditions, severe weight loss, and gut inflammation in the SARS-CoV-2 infected and BET inhibitor-treated K18- hACE2 mice model.
The findings are in line with a previous study that reported therapeutic application of BET inhibitor on K18-hACE2 mice resulted in severe lung pathology and significant viral RNA in the lung. The present study highlights the importance of BET proteins as positive regulators of antiviral gene expression.
Conclusion
The current study reveals the impact of the therapeutic application of the BET inhibitors during COVID-19 infection. Researchers have shed new light on the antiviral function of BET proteins during the infection. The authors advised against the use of BET inhibitors during SARS-CoV-2 infection as it could enhance the viral load.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Chen, I. et al. (2021) Viral E Protein Neutralizes BET Protein-Mediated Post-Entry Antagonism of SARS-CoV-2. bioRxiv 2021.11.14.468537; doi: https://doi.org/10.1101/2021.11.14.468537, https://www.biorxiv.org/content/10.1101/2021.11.14.468537v1
- Peer reviewed and published scientific report.
Chen, Irene A, James E Longbotham, Sarah McMahon, Rahul K Suryawanshi, Mir M Khalid, Taha E Taha, Takako Tabata, et al. 2022. “Viral E Protein Neutralizes BET Protein-Mediated Post-Entry Antagonism of SARS-CoV-2.” Cell Reports 40 (3): 111088–88. https://doi.org/10.1016/j.celrep.2022.111088. https://www.cell.com/cell-reports/fulltext/S2211-1247(22)00890-7?.