The majority of human infectious diseases are caused by pathogens that first spread in non-human animal species. "Zoonotic spillover" refers to the spread of infections from wild animals to humans.
Increased risk of spillover occurrences is linked to activities and variables that increase human connection with various animal species and the pathogens that they might harbor.
Over the last two decades, the ongoing coronavirus disease 2019 (COVID-19) is the third reported animal-to-human CoV spillover to have resulted in a severe epidemic. Coronavirus (CoV) zoonotic transmissions pose a significant threat to human health, with a large number of unknown reservoir hosts.
Alphacoronavirus (α) and betacoronavirus (β) are two serologic groups of coronaviruses that infect mammals. Notably, the Alphacoronavirus 1 infects dogs, cats, and pigs. Thus, the spike protein of these viruses plays a crucial role in the cellular entry.
Recently, a new study found Canine Coronavirus (CCoV) in nasopharyngeal swabs from a limited group of patients hospitalized with pneumonia in Sarawak, Malaysia, between 2017 and 2018. It was reported that CCoV-HuPn-2018 spike protein's N-terminus subdomain (0-domain) shares sequence similarities with Transmissible Gastroenteritis Virus (TGEV) and CCoV2b strains, but not with other members of type II Alphacoronaviruses.
 Study: Recent zoonotic spillover and tropism shift of a Canine Coronavirus is associated with relaxed selection and putative loss of function in NTD subdomain of spike protein. Image Credit: magic pictures / Shutterstock
Study: Recent zoonotic spillover and tropism shift of a Canine Coronavirus is associated with relaxed selection and putative loss of function in NTD subdomain of spike protein. Image Credit: magic pictures / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The Study
A new research paper published on the bioRxiv* preprint server describes selection pressures across the spike gene, concentrating on where such events are in connection to spike functional domains, and provides a new outlook on the recombination history involving the spike gene of CCoV-HuPn-2018 with other members of the Alphacoronavirus 1 type II species.
In this study, a genetic algorithm for recombination detection (GARD) was utilized to identify recombinant partitions in the N –terminal subdomains. The extensive analysis has led the authors to propose that CCoV-HuPn-2018 had an enteric origin but has since lost that tropism caused by mutations in the 0-domain's sialic acid binding site, resulting in lower selection pressure for this subdomain. These have evolved a respiratory tropism, similar to other Alphacoronaviruses 1 that have lost this area totally.
It was speculated that the virus might eventually lose this 0-domain region entirely and currently, we are experiencing the onset of this process.
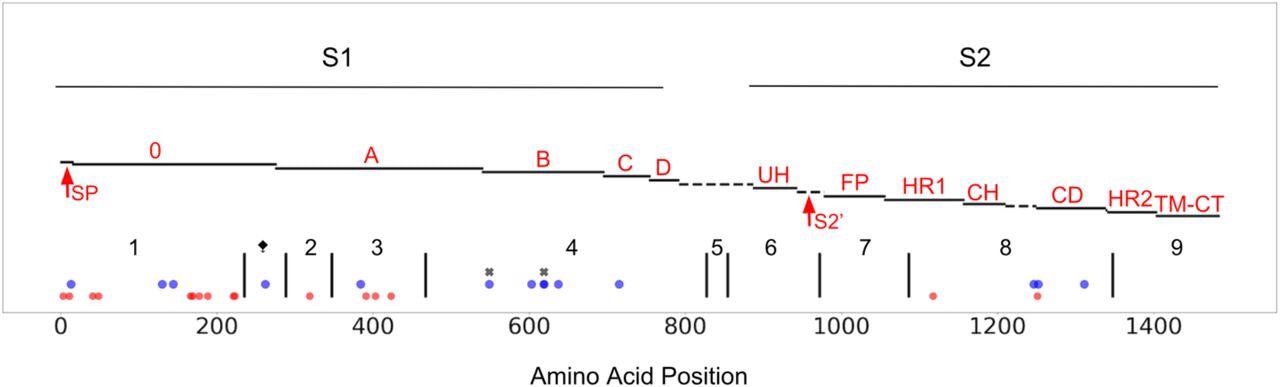 CCoV-HuPn-2018 spike protein, mapped to a published FIPV-UU4 spike gene map (Yang et al. 2020). S1, S2, of spike are highlighted and the protein is further subdivided into functional subunits and subdomains. Blue dots represent sites under positive selection in CCoV-HuPn-2018 as identified by MEME and FEL in the single branch tests; red dots represent sites that are unique in CCoV-HuPn-2018, but are not under positive selection; black “x”s indicate sites under positive selection in the MEME test of the complete alignment that had moderate EBF values for CCoV-HuPn-2018. Red text labels accompany each subdomain/functional unit and are based on the original FIPV spike structure (Yang et al. 2020): SP, signal peptide; 0 domain; A domain; B, includes RBD-Receptor-Binding Domain; C; D; UH, upstream helix; S2’, S2’ cleavage (predicted furin site, using ProP1.0 (Duckert et al. 2004)); FP, fusion peptide; HR1, heptad repeat region 1; CH, central helix; CD, connector domain; HR2, heptad repeat region 2; TM, transmembrane domain; CT, cytoplasmic tail. The dashed line between D and UH refers to a region of peptide with no sequence similarity between FIPV and CCoV-HuPn-2018; this region includes the S1/S2 furin cleavage site in FIPV, which is absent in CCoV-HuPn-2018. The vertical black lines represent the breakpoints of the GARD identified non-recombinant fragments, and are labeled numerically. The ⧪symbol represents a 3’ GARD fragment of alignment set I that was analyzed for positive selection; this GARD fragment was determined from an alignment of just CCoV2b and TGEV sequences (set I). The 5’ end of GARD fragment 2 represents the onset of FCoV2 sequence similarity (set II).
CCoV-HuPn-2018 spike protein, mapped to a published FIPV-UU4 spike gene map (Yang et al. 2020). S1, S2, of spike are highlighted and the protein is further subdivided into functional subunits and subdomains. Blue dots represent sites under positive selection in CCoV-HuPn-2018 as identified by MEME and FEL in the single branch tests; red dots represent sites that are unique in CCoV-HuPn-2018, but are not under positive selection; black “x”s indicate sites under positive selection in the MEME test of the complete alignment that had moderate EBF values for CCoV-HuPn-2018. Red text labels accompany each subdomain/functional unit and are based on the original FIPV spike structure (Yang et al. 2020): SP, signal peptide; 0 domain; A domain; B, includes RBD-Receptor-Binding Domain; C; D; UH, upstream helix; S2’, S2’ cleavage (predicted furin site, using ProP1.0 (Duckert et al. 2004)); FP, fusion peptide; HR1, heptad repeat region 1; CH, central helix; CD, connector domain; HR2, heptad repeat region 2; TM, transmembrane domain; CT, cytoplasmic tail. The dashed line between D and UH refers to a region of peptide with no sequence similarity between FIPV and CCoV-HuPn-2018; this region includes the S1/S2 furin cleavage site in FIPV, which is absent in CCoV-HuPn-2018. The vertical black lines represent the breakpoints of the GARD identified non-recombinant fragments, and are labeled numerically. The ⧪symbol represents a 3’ GARD fragment of alignment set I that was analyzed for positive selection; this GARD fragment was determined from an alignment of just CCoV2b and TGEV sequences (set I). The 5’ end of GARD fragment 2 represents the onset of FCoV2 sequence similarity (set II).
Findings
Four positively chosen sites were found within the putative receptor-binding domain (RBD) of CCoV-HuPn-2018. One of these positively selected sites is in a putative RBD extended loop, specifically near the end of extended loop 2, based on the feline infectious peritonitis virus (FIPV) spike structure and the concomitant CoV alignment.
In other Alphacoronaviruses, RBD extended loops establish contact locations with the aminopeptidase N (APN) receptor, and the specifics of the interaction between these loops and APN. Further, positive selection in CCoV-signal HuPn-2018's peptide could represent an adaptive role in this novel host, and the unique amino acid alterations in the signal peptide of CCoV-signal HuPn-2018's peptide compared to CCoV2b and TGEV could play a role in this strain's N-linked glycan repertoire.
It was hypothesized that the 0-domain of CCoV-spike HuPn-2018's protein might have lost its functional importance at some point in its history. Viruses such as porcine respiratory coronavirus (PRCV) have followed a similar evolutionary path, with the deletion of this portion of the protein being linked to a shift from enteric to respiratory tropism.
The high prevalence of Feline CoV2 (FCoV2) detected in Malaysian cats, concomitant with their primer construction strategy, suggests the possibility that an FCoV2 recombinant (WSU 79-1683)-like virus could be the prevalent FCoV2 strain in Malaysia. Previous experiments have demonstrated that feline APN can serve as a functional receptor of type II CCoV, TGEV and HCoV-229E. Hence, the infection in cats may be instrumental in generating recombinant Alphacoronavirus 1 CoVs. In conclusion, WSU 395 79-1683 or its close relative, has had a prominent role in the evolution of CCoV-HuPn-2018. These viruses have repeatedly coinfected hosts, leading to the development of recombinant progeny.
The molecular details of how the loss of this domain contributes to tropic shifts of this type, as well as the reason(s) why zoonotic host shifts in CoVs are generally associated with respiratory infection, remain unexplored mysteries.
The origins of CCoV-HuPn-2018, which date back to around 1957 suggest that this virus may have been circulating unnoticed for decades among humans, dogs, cats, and undiscovered intermediate hosts.
Therefore, the researchers entirely agree that a systematic survey for the prevalence of CCoV-HuPn-2018 in the host species—that make up the virus's complex history— should be conducted.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Zehr, J., Kosakovsky Pond, S., Martin, D., et al. (2021), Recent zoonotic spillover and tropism shift of a Canine Coronavirus is associated with relaxed selection and putative loss of function in NTD subdomain of spike protein, bioRxiv, doi: 10.1101/2021.11.15.468709, https://www.biorxiv.org/content/10.1101/2021.11.15.468709v1
- Peer reviewed and published scientific report.
Zehr, Jordan D., Sergei L. Kosakovsky Pond, Darren P. Martin, Kristina Ceres, Gary R. Whittaker, Jean K. Millet, Laura B. Goodman, and Michael J. Stanhope. 2022. “Recent Zoonotic Spillover and Tropism Shift of a Canine Coronavirus Is Associated with Relaxed Selection and Putative Loss of Function in NTD Subdomain of Spike Protein.” Viruses 14 (5): 853. https://doi.org/10.3390/v14050853. https://www.mdpi.com/1999-4915/14/5/853.