A team of scientists from the US has recently evaluated the susceptibility of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in sheep. The findings reveal that sheep are generally less susceptible to SARS-CoV-2 infection. However, the susceptibility is relatively higher for the alpha variant of the virus. The study is currently available on the bioRxiv* preprint server.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative pathogen of coronavirus disease 2019 (COVID-19), is an enveloped, positive-sense, single-stranded RNA virus that primarily affects epithelial cells of the human respiratory tract.
Being a zoonotic virus, it also affects a wide range of animals, serving as potential reservoirs to facilitate viral evolution and the emergence of novel variants.
According to available literature, more than 598 natural SARS-CoV-2 infections have been detected in 14 different animal species, including cats, dogs, large cats, gorillas, mink, and white-tailed deer. In addition, mice, hamsters, ferrets, and non-human primates have shown susceptibility to experimental SARS-CoV-2 infection.
The study
In the current study, the scientists have investigated whether sheep are susceptible to natural and experimental SARS-CoV-2 infection. Sheep have been chosen as an experimental model as they are often maintained in large groups and interact with humans during standard farming processes.
Both in vivo and in vitro studies were conducted. In the in vitro part, ruminant-derived cell lines were infected with SARS-CoV-2, and the viral growth was assessed. In the in vivo part, eight sheep were challenged with a mixture of original SARS-CoV-2 strain and alpha variant (B.1.1.7) via intranasal and oral routes simultaneously. After one day of the challenge, two naïve (uninfected) sheep were co-mingled with the infected sheep to study animal-to-animal viral transmission.
In a separate set of experiments, the competition between the original viral strain and alpha variant to infect sheep was assessed. This experiment could provide valuable information regarding the role of sheep in SARS-CoV-2 ecology.
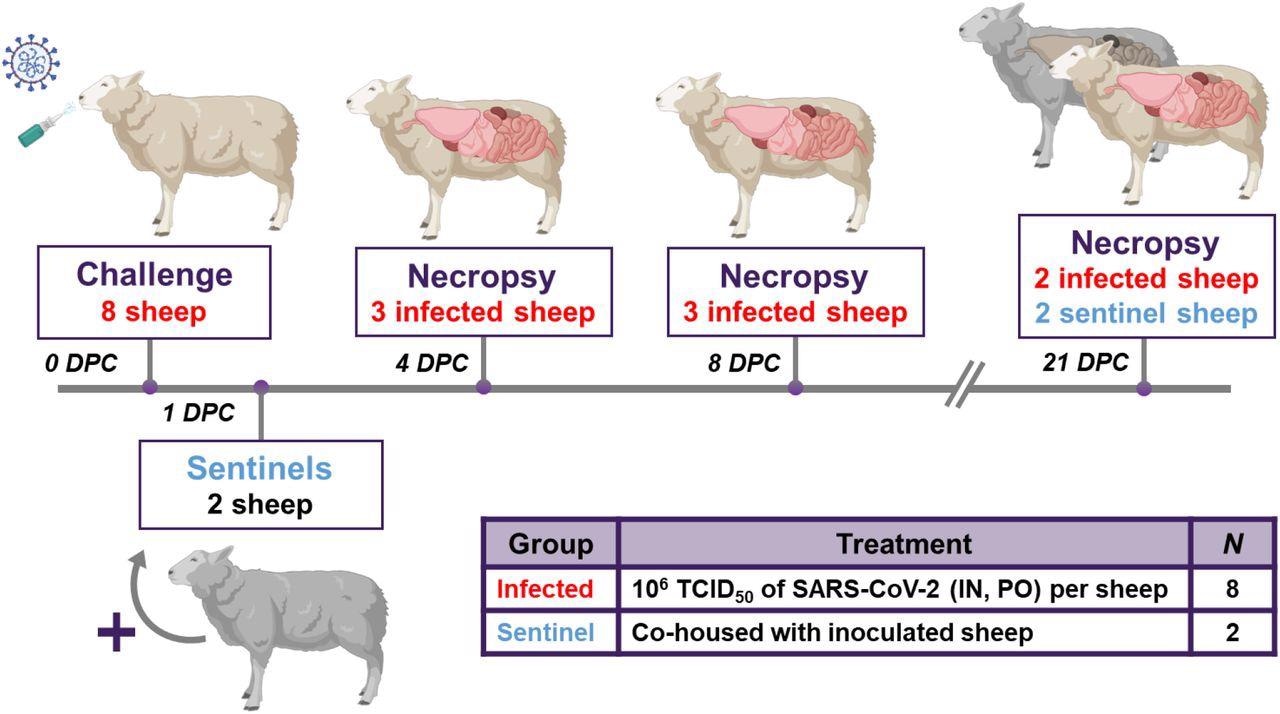 Eight sheep were inoculated with a mixture of SARS-CoV-2/human/USA/WA1/2020 lineage A (referred to as lineage A WA1; BEI item #: NR-52281) and SARS-CoV-2/human/USA/CA_CDC_5574/2020 lineage B.1.1.7 (alpha VOC B.1.1.7; NR-54011) acquired from BEI Resources (Manassas, VA, USA). A 2ml dose of 1×106 TCID50 per animal was administered IN and PO. At 1-day post challenge (DPC), 2 sentinel sheep were co-mingled with the 8 principal infected animals to study virus transmission. Daily clinical observations and body temperatures were performed. Nasal/oropharyngeal/rectal swabs, blood/serum and feces were collected at 0, 1, 3, 5, 7, 10, 14 17 and 21 DPC. Postmortem examinations were performed at 4 (3 principal), 8 (3 principal) and 21 DPC (2 principal + 2 sentinels). BioRender.com was used to create figure illustrations.
Eight sheep were inoculated with a mixture of SARS-CoV-2/human/USA/WA1/2020 lineage A (referred to as lineage A WA1; BEI item #: NR-52281) and SARS-CoV-2/human/USA/CA_CDC_5574/2020 lineage B.1.1.7 (alpha VOC B.1.1.7; NR-54011) acquired from BEI Resources (Manassas, VA, USA). A 2ml dose of 1×106 TCID50 per animal was administered IN and PO. At 1-day post challenge (DPC), 2 sentinel sheep were co-mingled with the 8 principal infected animals to study virus transmission. Daily clinical observations and body temperatures were performed. Nasal/oropharyngeal/rectal swabs, blood/serum and feces were collected at 0, 1, 3, 5, 7, 10, 14 17 and 21 DPC. Postmortem examinations were performed at 4 (3 principal), 8 (3 principal) and 21 DPC (2 principal + 2 sentinels). BioRender.com was used to create figure illustrations.
SARS-CoV-2 replication in ruminant-derived cells
The cells derived from sheep, cattle, and American pronghorn were analyzed for SARS-CoV-2 susceptibility. The findings revealed that both primary and immortalized sheep-derived kidney cells were susceptible to viral infection and that viral titer increased with time post-infection. In contrast, no viral replication was observed in cells derived from cattle and pronghorn.
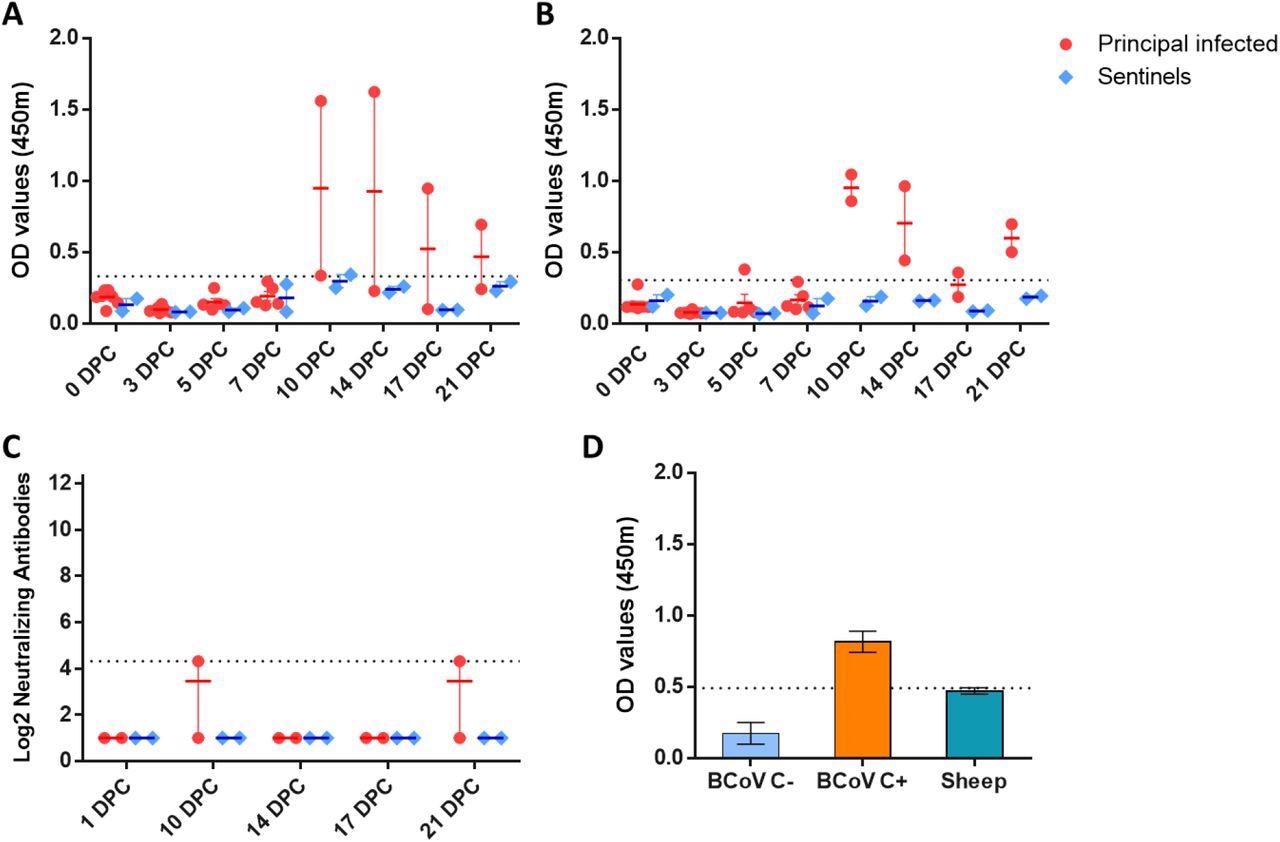 Serology of SARS-CoV-2 infected sheep. Detection of SARS-CoV-2 nucleocapsid protein (A), and the receptor binding domain (B) by indirect ELISA tests. The cut-off was determined by averaging the OD of negative serum + 3X the standard deviation as indicated by the dotted line. All samples with resulting OD values above this cut-off were considered positive. (C) Virus neutralizing antibodies detected in serum are shown as log2 of the reciprocal of the neutralization serum dilution. Sera were tested starting at a dilution of 1:20 which is indicated by the dotted line. (D) Sera from principal infected (n=8) and sentinel sheep (n=2) were tested against the bovine coronavirus (BCoV) spike protein using an indirect ELISA; both, positive (C+) and negative (C-) bovine control sera were included. The cut-off was determined by averaging the OD of negative serum + 3X the standard deviation as indicated by the dotted line. A-D: Mean with SEM are shown.
Serology of SARS-CoV-2 infected sheep. Detection of SARS-CoV-2 nucleocapsid protein (A), and the receptor binding domain (B) by indirect ELISA tests. The cut-off was determined by averaging the OD of negative serum + 3X the standard deviation as indicated by the dotted line. All samples with resulting OD values above this cut-off were considered positive. (C) Virus neutralizing antibodies detected in serum are shown as log2 of the reciprocal of the neutralization serum dilution. Sera were tested starting at a dilution of 1:20 which is indicated by the dotted line. (D) Sera from principal infected (n=8) and sentinel sheep (n=2) were tested against the bovine coronavirus (BCoV) spike protein using an indirect ELISA; both, positive (C+) and negative (C-) bovine control sera were included. The cut-off was determined by averaging the OD of negative serum + 3X the standard deviation as indicated by the dotted line. A-D: Mean with SEM are shown.
SARS-CoV-2 infection in sheep
The Sheep infected with the original virus and its alpha variant was studied along with their two contacts (naïve sheep) over a period of 21 days post-challenge. The findings revealed that none of the animals develop fever or any other clinical symptoms (weight loss, lethargy, diarrhea, or breathing difficulty) during the entire study period.
The presence of viral RNA and infectious virus was determined using nasal, oral, and rectal samples collected from infected animals and their contacts. Viral RNA was detected in nasal samples collected from infected sheep at day 1 post-challenge. No viral RNA was detected in rectal samples. Similarly, infectious virus was absent in all samples.
No viral RNA or infectious virus was detected in samples collected from two naïve animals during the 21-day study period.
In respiratory tract tissues collected from infected animals, viral RNA was detected at day 4 post-challenge, which remained detectable up to day 21. The highest amount of viral RNA was detected in the trachea, followed by the nasopharynx.
The lymphoid tissues collected from infected animals showed the presence of viral RNA at days 4, 8, and 21 post-challenge. The highest amount of RNA was detected in the tonsil, and in the retropharyngeal and tracheobronchial lymph nodes.
At day 21 post-challenge, respiratory tissues collected from the contact sheep also exhibited detectable viral RNA. Similarly, one contact animal showed the presence of viral RNA in the lymphoid tissue.
Regarding COVID-19-related pathologies, no significant pulmonary lesion was observed in infected animals. The viral antigen detected in the respiratory tract was solely associated with lymphoid aggregates observed in infected animals' upper and lower respiratory tracts.
Competition between original SARS-CoV-2 strain and alpha variant
The respiratory samples collected from co-infected sheep were analyzed by next-generation sequencing to determine the presence of each viral strain.
The findings revealed that the alpha variant of SARS-CoV-2 significantly replaced the original viral strain in sheep. However, while 100% presence of the alpha variant was detected in almost all samples tested, a very low percentage of original strain was detected only in some of the tested tissues.
Study significance
The study findings reveal that sheep are less susceptible to SARS-CoV-2 and develop only subclinical infection upon exposure. Although infected sheep are able to transmit the virus to other co-mingled sheep, the possibility of developing a productive secondary infection is very low.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Gaudreault NN. 2021. Susceptibility of sheep to experimental co-infection with the ancestral lineage of SARS-CoV-2 and its alpha variant, bioRxiv, https://www.biorxiv.org/content/10.1101/2021.11.15.468720v1
- Peer reviewed and published scientific report.
Gaudreault, Natasha N., Konner Cool, Jessie D. Trujillo, Igor Morozov, David A. Meekins, Chester McDowell, Dashzeveg Bold, et al. 2022. “Susceptibility of Sheep to Experimental Co-Infection with the Ancestral Lineage of SARS-CoV-2 and Its Alpha Variant.” Emerging Microbes & Infections 11 (1): 662–75. https://doi.org/10.1080/22221751.2022.2037397. https://www.tandfonline.com/doi/full/10.1080/22221751.2022.2037397.