The causative agent responsible for the unprecedented coronavirus disease 2019 (COVID-19) pandemic is severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
The mechanism of entry of SARS-CoV-2 involves the binding of the viral spike (S) protein to the cellular angiotensin-converting enzyme (ACE) 2 receptor that leads to proteolytic processing of the S precursor into the active S1 and S2 subunits. However, there are other factors responsible for SARS-CoV-2 entry, propagation, and pathogenesis.
In a previous study, researchers have shown that a variant of SARS-CoV-2 isolated in the Netherlands on 12 February 2020 seizes interferon-induced transmembrane (IFITM) proteins, especially IFITM2, as entry cofactors during infection.
The World Health Organization (WHO) has classified four SARS-CoV-2 variants as variants of concern (VOCs): B.1.1.7, B.1.351, P.1, and B.1.617.2. These are also known as alpha, beta, gamma, and delta variants, respectively.
In a study recently published on the bioRxiv* preprint server, researchers investigated the IFITM2 dependency of SARS-CoV-2 VOCs, including the delta variant for efficient infection.
 Study: FITM dependency of SARS-CoV-2 variants of concern. Image Credit: Pong Ch
Study: FITM dependency of SARS-CoV-2 variants of concern. Image Credit: Pong Ch

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Interferon-induced transmembrane proteins for efficient SARS-CoV-2 infection
IFITMs are a family of IFN stimulated genes (ISGs) known to protect cells against infection by many viruses, such as human immunodeficiency viruses, influenza A, rhabdo, and highly pathogenic coronaviruses, including SARS-CoV-2.
In this study, the researchers used the human epithelial lung cancer cell line Calu-3 due to its ability for siRNA knockdown (KD) of IFITM expression and iPSC-derived alveolar epithelial type II (iATII) cells as a model for the primary target cells of SARS-CoV-2 infection in the distal lung.
The Western blot analysis could not detect the ACE2 expression by iATII cells; however, it was easily detectable using fluorescence-activated cell sorting.
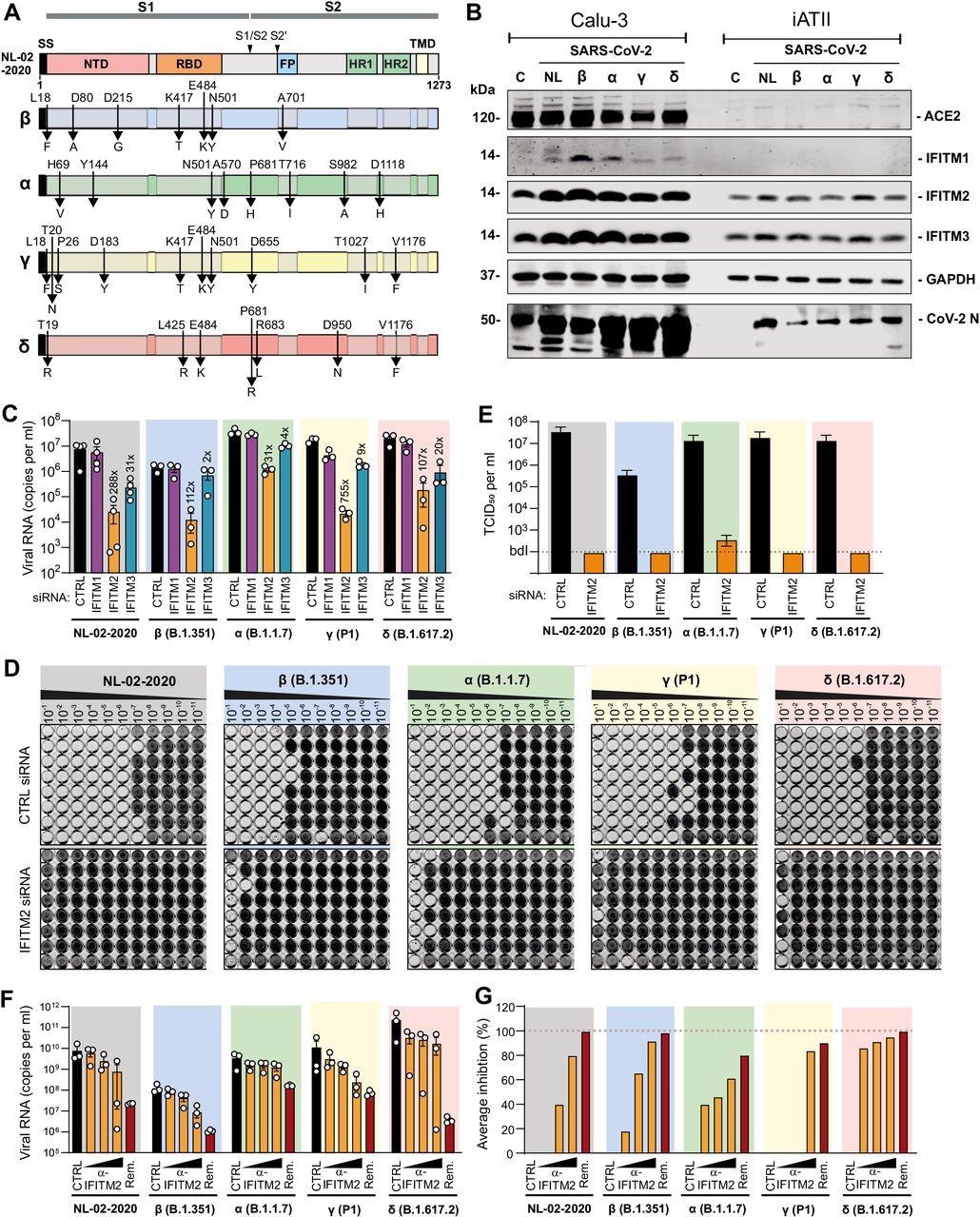 Effects of IFITM proteins on replication of SARS-CoV-2 variants. (A) Mutations in the Spike proteins of SARS-CoV-2 VOCs (Beta, blue; Alpha, green; Gamma, yellow; Delta, red) compared to the SARS-CoV-2 (NL-02-2020) strain. (B) Immunoblot of Calu-3 and iATII cells left uninfected (c) or infected with the indicated SARS-CoV-2 variants. Whole-cell lysates were stained with the indicated antibodies. An unspecific signal was observed in the Calu-3 control lane stained with the CoV-2 N antibody. (C) Viral N RNA levels in the supernatant of Calu-3 cells, collected 48 h post-infection with SARS-CoV-2 (MOI 0.05). Cells were transfected with control (CTRL) or IFITM targeting siRNAs as indicated. Bars represent the mean of 3 to 4 independent experiments (±SEM). (D) Impact of IFITM2 siRNA knock-down on infectious SARS-CoV-2 yields. Supernatants derived from Calu-3 cells treated with control (CTRL) of IFITM2 siRNA two days after infection with the SARS-CoV-2 NL-02-2020 or VOCs were serially diluted and added to Caco-2 cells seeded in 96-well plates. Five days later cells were examined for CPE, fixed and stained with crystal violet. Productively infected wells appear transparent since cells are eliminated and detached by the virus. (E) Infectious SARS-CoV-2 particles in the supernatant of Calu-3 cells treated with control or IFITM2 targeting siRNAs. Bars represent the mean of one experiment performed with eight technical replicates (±SD) shown in panel D. (F) Quantification of viral N RNA levels in the supernatant of iATII cells treated with α-IFITM2 antibody (20, 40, or 80 μg/ml) or Remdesivir (10 μM) 1 h before infection (SARS-CoV-2, MOI 0.5), collected 48 h post-infection. Bars represent the mean of three independent experiments. (G) Average percentage of reduction of vRNA levels in the supernatants of (E) compared to the untreated control.
Effects of IFITM proteins on replication of SARS-CoV-2 variants. (A) Mutations in the Spike proteins of SARS-CoV-2 VOCs (Beta, blue; Alpha, green; Gamma, yellow; Delta, red) compared to the SARS-CoV-2 (NL-02-2020) strain. (B) Immunoblot of Calu-3 and iATII cells left uninfected (c) or infected with the indicated SARS-CoV-2 variants. Whole-cell lysates were stained with the indicated antibodies. An unspecific signal was observed in the Calu-3 control lane stained with the CoV-2 N antibody. (C) Viral N RNA levels in the supernatant of Calu-3 cells, collected 48 h post-infection with SARS-CoV-2 (MOI 0.05). Cells were transfected with control (CTRL) or IFITM targeting siRNAs as indicated. Bars represent the mean of 3 to 4 independent experiments (±SEM). (D) Impact of IFITM2 siRNA knock-down on infectious SARS-CoV-2 yields. Supernatants derived from Calu-3 cells treated with control (CTRL) of IFITM2 siRNA two days after infection with the SARS-CoV-2 NL-02-2020 or VOCs were serially diluted and added to Caco-2 cells seeded in 96-well plates. Five days later cells were examined for CPE, fixed and stained with crystal violet. Productively infected wells appear transparent since cells are eliminated and detached by the virus. (E) Infectious SARS-CoV-2 particles in the supernatant of Calu-3 cells treated with control or IFITM2 targeting siRNAs. Bars represent the mean of one experiment performed with eight technical replicates (±SD) shown in panel D. (F) Quantification of viral N RNA levels in the supernatant of iATII cells treated with α-IFITM2 antibody (20, 40, or 80 μg/ml) or Remdesivir (10 μM) 1 h before infection (SARS-CoV-2, MOI 0.5), collected 48 h post-infection. Bars represent the mean of three independent experiments. (G) Average percentage of reduction of vRNA levels in the supernatants of (E) compared to the untreated control.
Outcomes of the study
The results showed that the SARS-CoV-2 Delta variant of concern in comparison to NL-02-2020 in Calu-3 cells generated 3-fold higher levels of viral RNA.
The outcomes of the study showed that there was a reduction of viral RNA production from 31- (Alpha) to 754-fold (Gamma) on depletion of endogenous IFITM2 expression in Calu-3 cells.
Further, it was noted that KD of IFITM1 had a small effect; however, silencing of IFITM3 resulted in 2- to 31-fold depletion of viral RNA production.
Interestingly, the production of infectious SARS-CoV-2 particles in Calu-3 cells reduced to near background levels upon silencing of IFITM2. Furthermore, the replication of SARS-CoV-2 VOC in iATII was reduced by the antibody targeting the N-terminus of IFITM2 in a dose-dependent manner.
"IFITMs (especially IFITM2) are also critical cofactors for efficient replication of current SARS-CoV-2 VOCs including the dominant Delta variant."
Conclusions
The study's findings suggest that endogenous IFITMs (especially IFITM2) are important cofactors for the effective replication of SARS-CoV-2 VOCs, including the dominant Delta variant. It was noted that the SARS-CoV-2 Delta variant replicated at a 30-times higher rate in comparison to the early NL-02-2020 isolate in iATII cells. Thus, the dependency of IFITM2 maintained by VOCs can be further explored as a target for therapeutic or preventive approaches.
Efficient inhibition of the SARS-CoV-2 Delta VOC by an a-IFITM2 antibody illustrates that IFITM2 may have a crucial role in SARS-CoV-2 transmission as well as pathogenesis.
"Our observation that IFITM2 dependency is maintained by VOCs also further underlines that this cellular "antiviral" factor represents an interesting target for therapeutic or preventive approaches."

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
IFITM dependency of SARS-CoV-2 variants of concern. Rayhane Nchioua, Annika Schundner, Dorota Kmiec, Caterina Prelli Bozzo, Fabian Zech, Lennart Koepke, Manfred Frick, Konstantin M. J. Sparrer, Frank Kirchhoff, bioRxiv, 2021. doi: https://doi.org/10.1101/2021.11.17.468942, https://www.biorxiv.org/content/10.1101/2021.11.17.468942v1
- Peer reviewed and published scientific report.
Rayhane Nchioua, Annika Schundner, Dorota Kmiec, Caterina Prelli Bozzo, Fabian Zech, Lennart Koepke, Alexander Graf, et al. 2022. “SARS-CoV-2 Variants of Concern Hijack IFITM2 for Efficient Replication in Human Lung Cells.” Journal of Virology 96 (11). https://doi.org/10.1128/jvi.00594-22. https://journals.asm.org/doi/10.1128/jvi.00594-22#.