Since the beginning of the coronavirus disease 2019 (COVID-19) pandemic, scientists across the world have been working tremendously hard to contain the pandemic. COVID-19 has been caused by the rapid spread of a novel coronavirus, namely, severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), which was first reported in 2019 in Wuhan, China. The scientific efforts have led to the generation of an extraordinary amount of publicly available data associated with the human immune response to SARS-CoV-2.
 Study: Characterization of SARS-CoV-2 public CD4+ αβ T cell clonotypes through reverse epitope discovery
Study: Characterization of SARS-CoV-2 public CD4+ αβ T cell clonotypes through reverse epitope discovery

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Importance of T Cell Receptor (TCR) Repertoire
Previous studies have emphasized the importance of T cell immune response to SARS-CoV-2 infection as it provides long-term protection. TCR repertoire sequencing is one of the popular methods used to analyze the complexity of the T cell response. TCR is a heterodimer of alpha and beta chains formed in a semi-random DNA recombination process, causing the generation of their unique repertoire in each individual. However, in certain situations, an individual’s T cells can represent similar features on encountering the same antigens. For instance, immunodominant epitopes induce significant clonal expressions, and when TCRs recognize such epitopes, they produce highly similar sequences.
Scientists believe that assessing TCR repertoires could help understand the difference and commonalities of the T cell immune response in every individual against the same antigens. This analysis could help identify targets for the development of therapeutics (e.g., adoptive T cell transfers) as well as diagnostic tools.
Recently, a COVID-19 diagnostic test has been developed based on the identification of T cell clonotypes reactive to SARS-CoV-2 antigens, which has received emergency use authorization from the US Food and Drug Administration (FDA).
Methods to Identify TCR Repertoire Signatures
Several studies are available that are associated with the identification of TCR repertoire signatures related to SARS-CoV-2 infection. Some have used bulk TCR repertoire sequencing to quantify the large numbers of unpaired TCRalpha or TCRbeta clonotypes. Additionally, scientists used single-cell TCR sequencing techniques that generated fewer but paired alpha/beta TCR sequences. Some researchers have also adopted T cell antigen-enrichment methodologies, e.g., MHC-multimer-staining or peptide stimulation with subsequent enrichment for activated cells to detect more SARS-CoV-2 specific TCRs in each sample, which would enable identification of immunodominant epitopes. One of the standard methods used to discover SARS-CoV-2 epitope is stimulating T cells with peptide libraries.
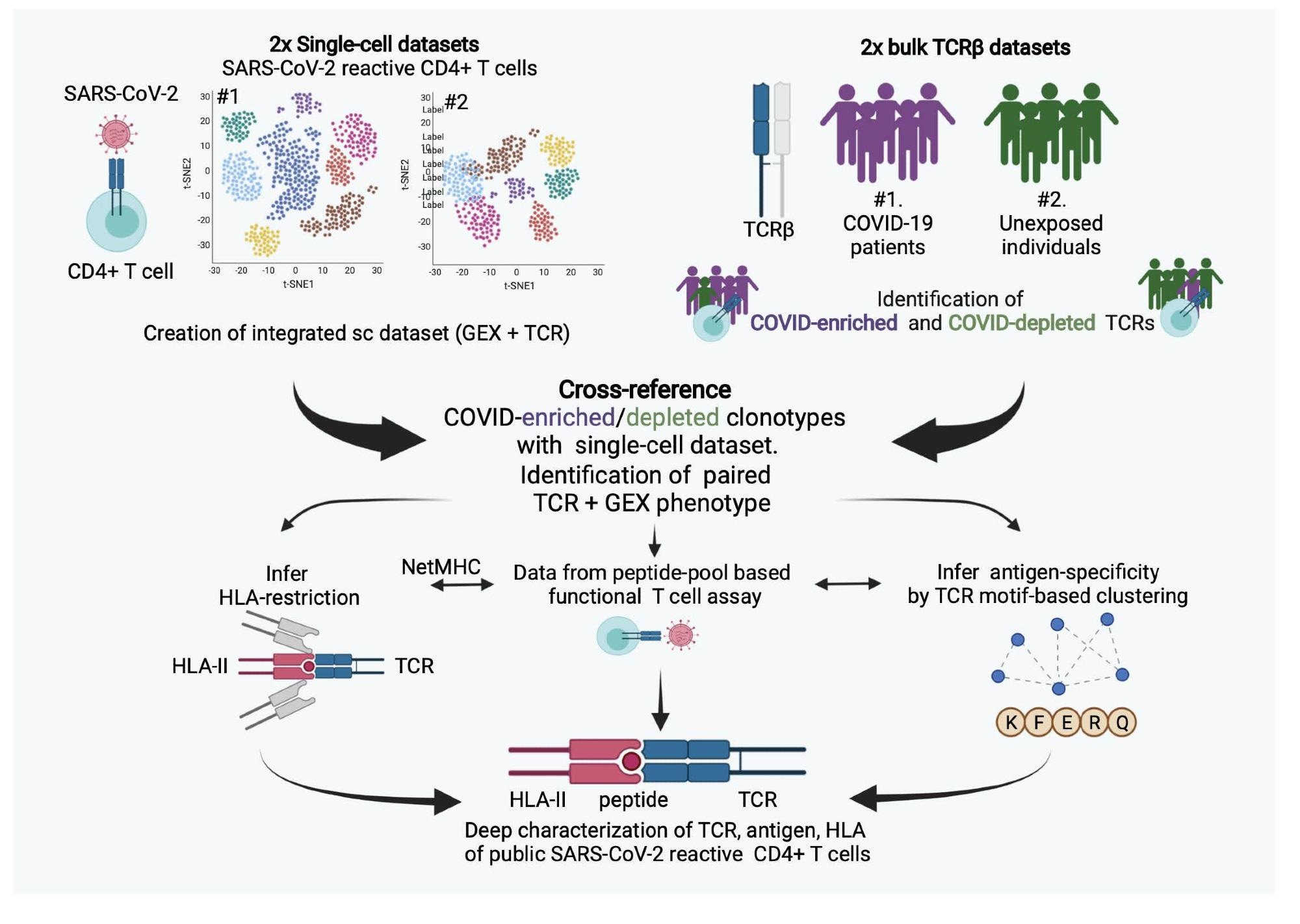
A new study, published on the bioRxiv* preprint server, has focused on public clonotypes, i.e., clones found in multiple individuals, for the development of a novel reverse epitope discovery technique. This technique holds greater potential for population-wide applications.
Scientists revealed that the reverse epitope discovery technique uses TCR repertoires to find immunodominant responses in an unbiased manner. They conducted a comprehensive TCR meta-analysis of publicly available single-cell and bulk TCR repertoire datasets and were able to find SARS-CoV-2 reactive TCRs with complete TCR alpha and beta chain information and inferred their human leukocyte antigen (HLA) restriction. Researchers used TCR clustering based on sequence similarity to identify several prominent alpha/beta TCR motifs as well as predict their antigen specificity.
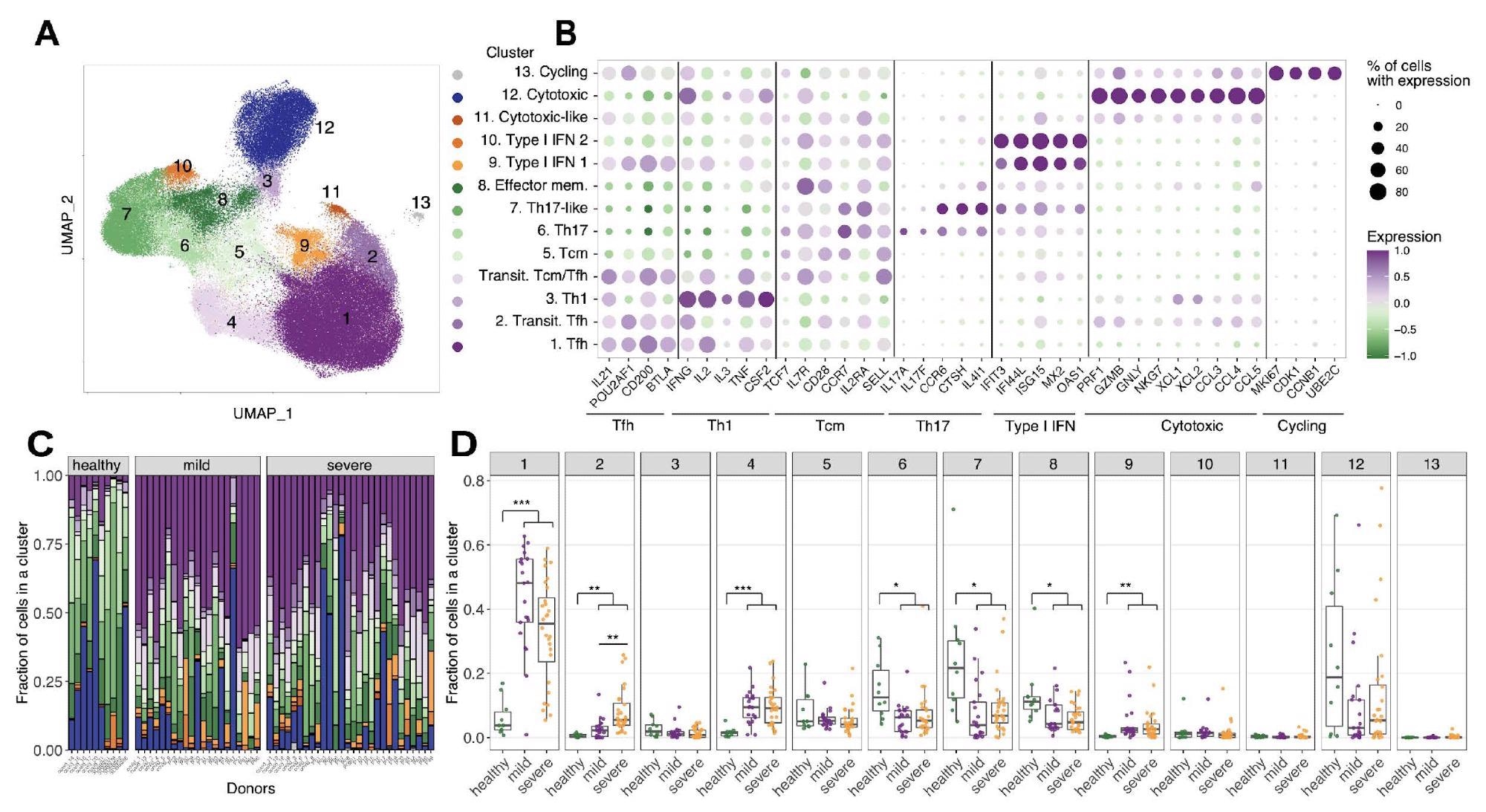 A. UMAP of single cells for merged (Bacher et al., 2020; Meckiff et al., 2020) datasets of SARS-CoV-2 antigen-enriched CD4 T cells based. Colors show clusters of cells with distinct gene expression profiles B. Differentially expressed genes in each GEX cluster. C. Distribution of cells between GEX clusters is plotted for each donor. Healthy donors less Tfh cells (populations 1 and 2) D. Boxplots showing cell proportion distribution among functional clusters for each patient (Mann-Whitney U test, Bonferroni multiple comparison correction).
A. UMAP of single cells for merged (Bacher et al., 2020; Meckiff et al., 2020) datasets of SARS-CoV-2 antigen-enriched CD4 T cells based. Colors show clusters of cells with distinct gene expression profiles B. Differentially expressed genes in each GEX cluster. C. Distribution of cells between GEX clusters is plotted for each donor. Healthy donors less Tfh cells (populations 1 and 2) D. Boxplots showing cell proportion distribution among functional clusters for each patient (Mann-Whitney U test, Bonferroni multiple comparison correction).
Study Implications
To find public CD4+ T cell responses to SARS-CoV-2 infection, the authors merged two publicly available single-cell datasets of CD4+ SARS-CoV-2-reactive T cells. This combined dataset comprised of 125,258 cells that have successfully undergone quality control steps and resulted in 13 functional clusters after unsupervised analysis.
Importantly, this study found that clusters were associated with helper follicular T (TfH) cells, helper type 1 T (Th1) cells, transitional Tfh/Tcm T cells, central memory T (Tcm) cells, Th17 phenotypes, effector memory T cells, type I IFN-signature T cells, cytotoxic T cells, and cycling T cells.
The authors reported that two main TfH T cell subsets were significantly different in hospitalized versus non-hospitalized COVID-19 patients. The integrative meta-analysis was used to characterize the clonotypes. This study reported that CD4+ COVID-enriched clonotypes possess T follicular helper functional features. Further, on the depletion of clonotypes, COVID-19 infected individuals mostly had a central memory phenotype.
This study successfully identified 1248 paired TCR clonotypes that are highly specific to immunogenic epitopes of SARS-CoV-2. This study not only identified but also characterized antigen-specificity for 428 TCRs using the MIRA dataset. Scientists have also determined HLA restriction for most of these TCRs. TCR-HLA pairings were also validated via NetMHC binding predictions and after vaccination with ChAdOx1. This study reported the presence of at least 20 unique COVID-enriched TCR sequences in every individual. Interestingly, this study reported that CD4-associated TCR repertoire differentiates vaccination from natural infection.
Conclusions
The authors mentioned that their method of reverse epitope discovery bears certain limitations. For instance, this method is strongly associated with public T cell responses. However, immunodominant responses are also caused by private clonotypes and these could not be identified by this method.
Another limitation includes the dependence on the availability of TCR antigen-specificity data from public databases and resources such as VDJdb and the MIRA dataset.
The main strength of this approach is the identification of both cross-reactive and immunodominant responses to SARS-CoV-2. In addition, researchers suggested that the wide-ranging coverage of HLA haplotypes presented in this study could help further investigation related to immunodominant responses in a genetically diverse population.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Rosati, E. et al. (2021) Characterization of SARS-CoV-2 public CD4+ αβ T cell clonotypes through reverse epitope discovery, bioRxiv, 2021.11.19.469229; doi: https://doi.org/10.1101/2021.11.19.469229, https://www.biorxiv.org/content/10.1101/2021.11.19.469229v1
- Peer reviewed and published scientific report.
Pogorelyy, Mikhail V., Elisa Rosati, Anastasia A. Minervina, Robert C. Mettelman, Alexander Scheffold, Andre Franke, Petra Bacher, and Paul G. Thomas. 2022. “Resolving SARS-CoV-2 CD4+ T Cell Specificity via Reverse Epitope Discovery.” Cell Reports Medicine 3 (8): 100697. https://doi.org/10.1016/j.xcrm.2022.100697. https://www.cell.com/cell-reports-medicine/fulltext/S2666-3791(22)00233-6?.