Scientists revealed that the identification of duplex ribonucleic acids (RNAs) via cellular RNA sensors is critically important to understand hosts' response to infections. This is because the recognition of duplex RNAs triggers signaling cascades responsible for the production of interferon (IFN) and, subsequent, upregulation of several interferon-stimulated genes (ISGs). Hence, this molecular pathway serves as a potential target for therapeutic development for various viral diseases.
 Study: Self-assembling short immunostimulatory duplex RNAs with broad spectrum antiviral activity. Image Credit: ktsdesign / Shutterstock
Study: Self-assembling short immunostimulatory duplex RNAs with broad spectrum antiviral activity. Image Credit: ktsdesign / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
Previous studies have reported that duplex RNAs comprise various structural features recognized by the three cellular RNA sensors that induce an innate immune response. These cellular RNA sensors include toll-like receptor 3 (TLR3), a retinoic acid-inducible gene I (RIG-I), and melanoma differentiation-associated gene 5 (MDA5). TL3 is located on the cell membrane, and endosomal membrane and RIG-I and MDA5 are located in the cytosol.
Scientists revealed that long forms of duplex RNA are identified by these sensors based on their length. TLR3 senses duplex RNAs of more than 35 bp in length and MDA5 recognizes duplex RNAs that are above 300 bp in length, while a short stretch of duplex RNA (below 19 bp) is identified by RIG-I.
Previous studies revealed that duplex RNA-mediated innate immune responses are a two-edged sword. This is because, in the case of respiratory infections caused by viruses such as SARS-CoV-2, duplex RNA-induced innate immune responses provide the first line of host defense against the invading virus; however, utilization of duplex RNAs for RNA interference (RNAi) methods could cause unfavorable immunological off-target effects. In addition, this could enable misinterpretation of experimental results. Therefore, a better understanding of the mechanism related to cells' response to duplex RNAs could have a lasting impact on biology and medicine research.
A New Study
A new study, published on the bioRxiv* preprint server, has discovered a class of new immunostimulatory RNAs while studying influenza infection-associated host genes in human lung epithelial cells using small interfering RNAs (siRNAs). Scientists revealed that these short duplex RNAs could induce type I and type III interferons (IFN-I/III) in a wide variety of cells.
Systematic mechanistic analysis has shown that these immunostimulatory RNAs can activate the RIG-I/IRF3 pathway via directly attaching to RIG-I. This binding depends on a few factors such as the short RNAs comprising a conserved overhanging sequence motif and the minimum sequence length must be of 20 bases.
Researchers explained that the conserved overhanging motif is accountable for the self-assembly of end-to-end RNA dimers through Hoogsteen G-G base pairing. The current study is the first to report that Hoogsteen base pairing can lead to the generation of duplex RNAs that are highly effective RIG-I agonists.
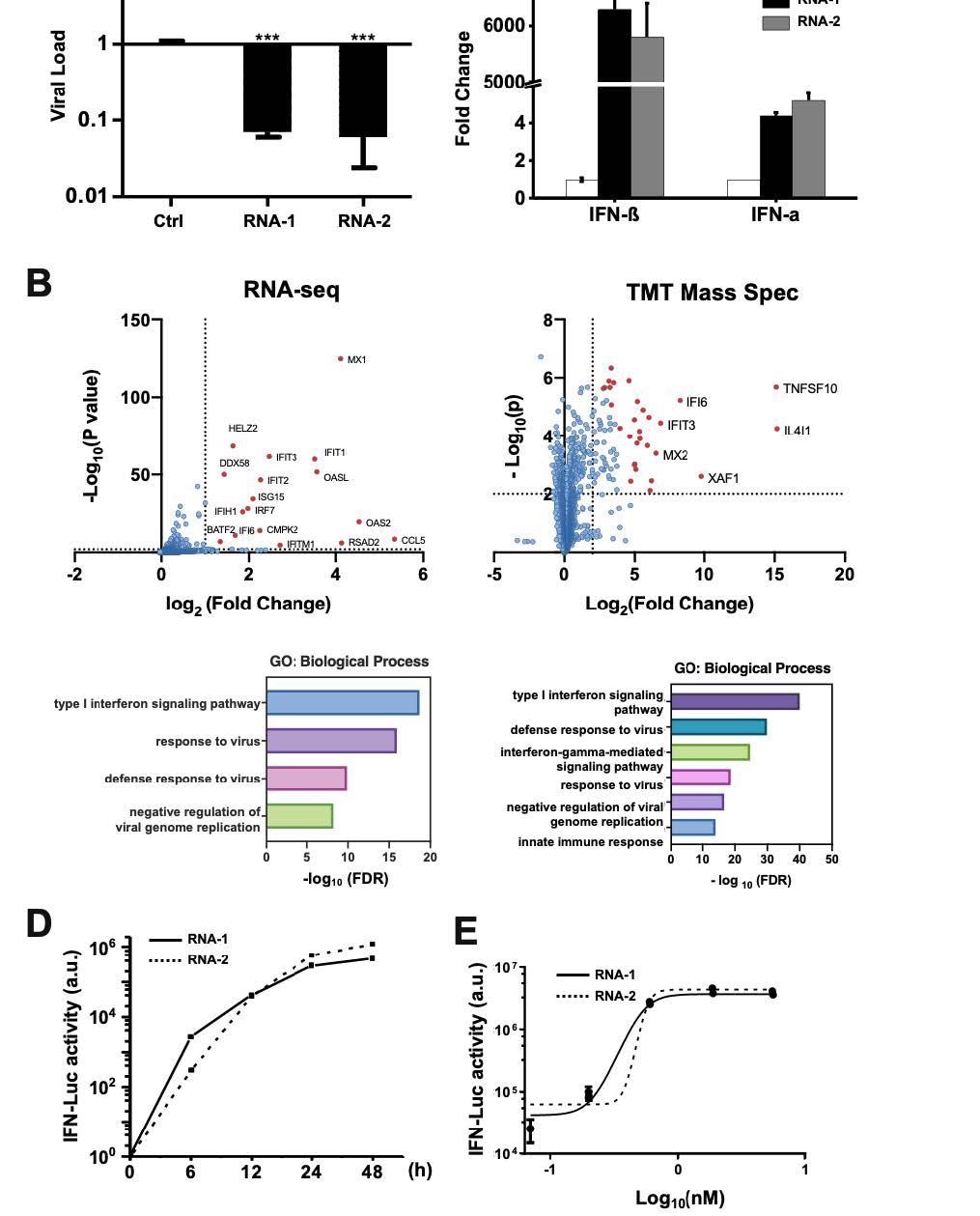 Discovery of new immunostimulatory RNAs. (A) A549 cells were transfected with RNA-1, RNA-2, or a scrambled duplex RNA control, and infected with influenza A/WSN/33 (H1N1) virus (MOI=0.01) 24 hours later. Titers of progeny viruses in medium supernatants collected at 48 h post-infection were determined by quantifying plaque-forming units (PFUs); data are shown as % viral infection measured in the cells treated with the control RNA (Data shown are mean ± standard deviation; N =3; ***, p < 0.001). (B) A549 cells were transfected with RNA-1 or a scrambled dsRNA control, collected at 48 h, and analyzed by RNA761 seq (left) or TMT Mass Spec (right). Differentially expressed genes (DEGs) from RNA-seq or proteins from TMT Mass Spec are shown in volcano plots (top) and results of GO Enrichment analysis performed for the DEGs are shown at the bottom (N = 3). (C) qPCR analysis of cellular IFN-β and IFN-α RNA levels at 48 h after A549 cells were transfected with RNA-1, RNA-2, or scrambled dsRNA control (N = 3). (D) RNA-mediated production kinetics of IFN production in wild-type A549-Dual cells that were transfected with RNA-1, RNA-2, or scramble RNA control measured using a Quanti-Luc assay. OD values from cells transfected with the scrambled RNA control were subtracted as background (N = 6). (E) Dose-dependent induction of IFN by RNA-1 and -2 in A549-Dual cells compared to scrambled RNA control measured at 48 h post770 transfection (control OD values were subtracted as background; N = 6).
Discovery of new immunostimulatory RNAs. (A) A549 cells were transfected with RNA-1, RNA-2, or a scrambled duplex RNA control, and infected with influenza A/WSN/33 (H1N1) virus (MOI=0.01) 24 hours later. Titers of progeny viruses in medium supernatants collected at 48 h post-infection were determined by quantifying plaque-forming units (PFUs); data are shown as % viral infection measured in the cells treated with the control RNA (Data shown are mean ± standard deviation; N =3; ***, p < 0.001). (B) A549 cells were transfected with RNA-1 or a scrambled dsRNA control, collected at 48 h, and analyzed by RNA761 seq (left) or TMT Mass Spec (right). Differentially expressed genes (DEGs) from RNA-seq or proteins from TMT Mass Spec are shown in volcano plots (top) and results of GO Enrichment analysis performed for the DEGs are shown at the bottom (N = 3). (C) qPCR analysis of cellular IFN-β and IFN-α RNA levels at 48 h after A549 cells were transfected with RNA-1, RNA-2, or scrambled dsRNA control (N = 3). (D) RNA-mediated production kinetics of IFN production in wild-type A549-Dual cells that were transfected with RNA-1, RNA-2, or scramble RNA control measured using a Quanti-Luc assay. OD values from cells transfected with the scrambled RNA control were subtracted as background (N = 6). (E) Dose-dependent induction of IFN by RNA-1 and -2 in A549-Dual cells compared to scrambled RNA control measured at 48 h post770 transfection (control OD values were subtracted as background; N = 6).
Additionally, the authors claimed that these immunostimulatory RNAs are novel as they are able to trigger the production of IFN irrespective of the presence of blunt or overhanging ends, terminal hydroxyl or monophosphate groups, and RNA base- or DNA base-ends.
This was not the case in the previously described immunostimulatory RNAs, as they showed definitive requirements such as 5’-di or triphosphates for the activation of cellular RNA sensors. These findings are in line with a previous study that revealed N1-2'O-methylation at the 5' end of the antisense strand, and the loss of the other ends of the original short dsRNA caused complete loss of the immunostimulatory activity.
The current study revealed that the RNA-mediated IFN-I/III production could significantly inhibit infection caused by many human respiratory viruses such as H1N1 and H3N2 influenza viruses, SARS-CoV-1, MERS-CoV, HCoV-NL, and SARS-CoV-2.
Importantly, these immunostimulatory RNAs can significantly decrease the SARS-CoV-2 viral loads in cell lines as well as in human lung airways and alveolus chips comprising primary lung epithelial and endothelial cells.
Researchers are optimistic that this study could aid in the development of siRNAs that could avoid undesired immune activation. Therefore, it could be used to design novel RNA therapeutics for both prevention and treatment of respiratory virus infections.
Conclusion
Some of the advantages of using the new duplex RNAs over the commonly used PRR agonist Poly(I:C) are that they can be easily synthesized and employ a more targeted antiviral effect with less proinflammatory activity.
The authors of this study revealed that the new immunostimulatory RNAs could specifically activate the RIG-I/IFN-I pathway and do not activate any other cellular RNA sensors, such as TLR7, TLR8, MDA5, or TLR3. They stated that if optimally designed, i.e., omitting the overhanging motif in siRNAs, unwanted activation of innate immune responses could be avoided. Additionally, the researchers emphasized that although stimulation of the immune system is not a desired phenomenon in some gene silencing applications, this application could be beneficial in others, such as the treatment of viral infection and cancer.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Si, L. et al. (2021) Self-assembling short immunostimulatory duplex RNAs with broad spectrum antiviral activity, bioRxiv, 2021.11.19.469183; doi: https://doi.org/10.1101/2021.11.19.469183, https://www.biorxiv.org/content/10.1101/2021.11.19.469183v1
- Peer reviewed and published scientific report.
Si, Longlong, Haiqing Bai, Crystal Yuri Oh, Amanda Jiang, Fan Hong, Tian Zhang, Yongxin Ye, et al. 2022. “Self-Assembling Short Immunostimulatory Duplex RNAs with Broad-Spectrum Antiviral Activity.” Molecular Therapy. Nucleic Acids 29 (August): 923–40. https://doi.org/10.1016/j.omtn.2022.08.031. https://www.cell.com/molecular-therapy-family/nucleic-acids/fulltext/S2162-2531(22)00230-X?.