Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first identified in Wuhan, China, in late December 2019, and subsequently led to the coronavirus disease 2019 (COVID-19) pandemic.
The SARS-CoV-2 infection is facilitated by the entry receptor angiotensin-converting enzyme 2 (ACE2). Attachment factors and co-receptors facilitating entry have been extensively studied. However, cellular entry factors inhibiting viral entry are largely unknown.
In a recent study, published on the bioRxiv* preprint server, researchers identified human LRRC15 as an inhibitory receptor for SARS-CoV-2 entry, which directly binds to the receptor-binding domain (RBD) of the spike protein with a moderate affinity and inhibits spike-mediated entry.
 Study: LRRC15 is an inhibitory receptor blocking SARS-CoV-2 spike-mediated entry in trans. Image Credit: Dotted Yeti / Shutterstock
Study: LRRC15 is an inhibitory receptor blocking SARS-CoV-2 spike-mediated entry in trans. Image Credit: Dotted Yeti / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background and Motivation
Similar to SARS-CoV-1, SARS-CoV-2 utilizes ACE2 as a primary entry receptor. The viral structural protein spike (S) binds to ACE2 and mediates the cellular entry of the virus. The S1 subunit consists of the N-terminal domain (NTD) and the receptor-binding domain (RBD).
The interaction between the RBD and ACE2 is important to determine several critical features of SARS-CoV-2 infection. The Spike protein (specifically the RBD) is the main target antigen for COVID-19 vaccines, which interfere with the binding between RBD and ACE2. This highlights the importance of RBD and its binding to the cellular receptor for controlling SARS-CoV-2.
Scientists have identified several cellular factors that facilitate cellular entry of SARS-CoV-2, but it is not clear if any cellular receptors inhibit viral entry. In the current study, researchers generated a focused CRISPRa library named surfaceome. The library covers approximately 6,000 of all known/predicted surface proteins on the cellular plasma membrane.
The screening method used revealed the human LRRC15 (leucine-rich repeat-containing 15) to be a novel inhibitory receptor for SARS-CoV-2.
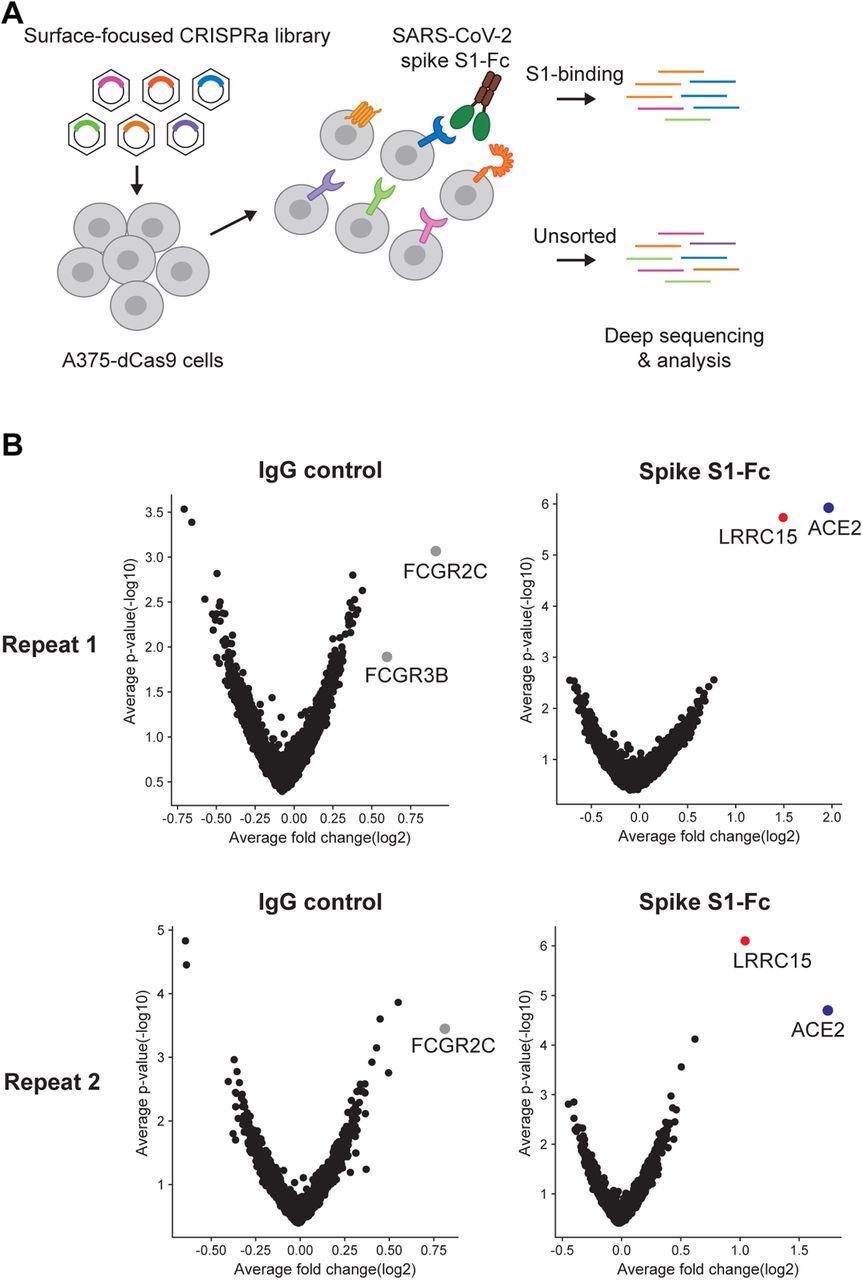 A surfaceome-focused CRISPR activation screen identified cellular receptors binding with SARS-CoV-2 spike protein (A) Schematic of a focused CRISPR activation screen for surface proteins interacting with SARS-CoV-2 spike S1-Fc fusion protein. (B) Volcano plots showing sgRNAs enriched or depleted in cells binding with SARS-CoV-2 spike S1-Fc or human IgG isotype control. Results from two biologically independent replicates are shown.
A surfaceome-focused CRISPR activation screen identified cellular receptors binding with SARS-CoV-2 spike protein (A) Schematic of a focused CRISPR activation screen for surface proteins interacting with SARS-CoV-2 spike S1-Fc fusion protein. (B) Volcano plots showing sgRNAs enriched or depleted in cells binding with SARS-CoV-2 spike S1-Fc or human IgG isotype control. Results from two biologically independent replicates are shown.
Key Findings
Scientists tried to identify cellular receptors for SARS-CoV-2 by staining cells with a recombinant spike protein, which produced two distinct hits, namely, the ACE2 and LRRC15. ACE2 is, of course, the bonafide entry receptor, while LRRC15 is the novel inhibitory receptor. Furthermore, it was observed that LRRC15 directly interacts with the RBD of S with moderate affinity.
It is known from previous research that ACE2 also interacts with S (through the RBD), but the LRRC15-RBD interaction was not found to compete with the ACE2-RBD interaction or stabilize it. However, more high-resolution studies will be needed to determine the interface of LRRC15-RBD.
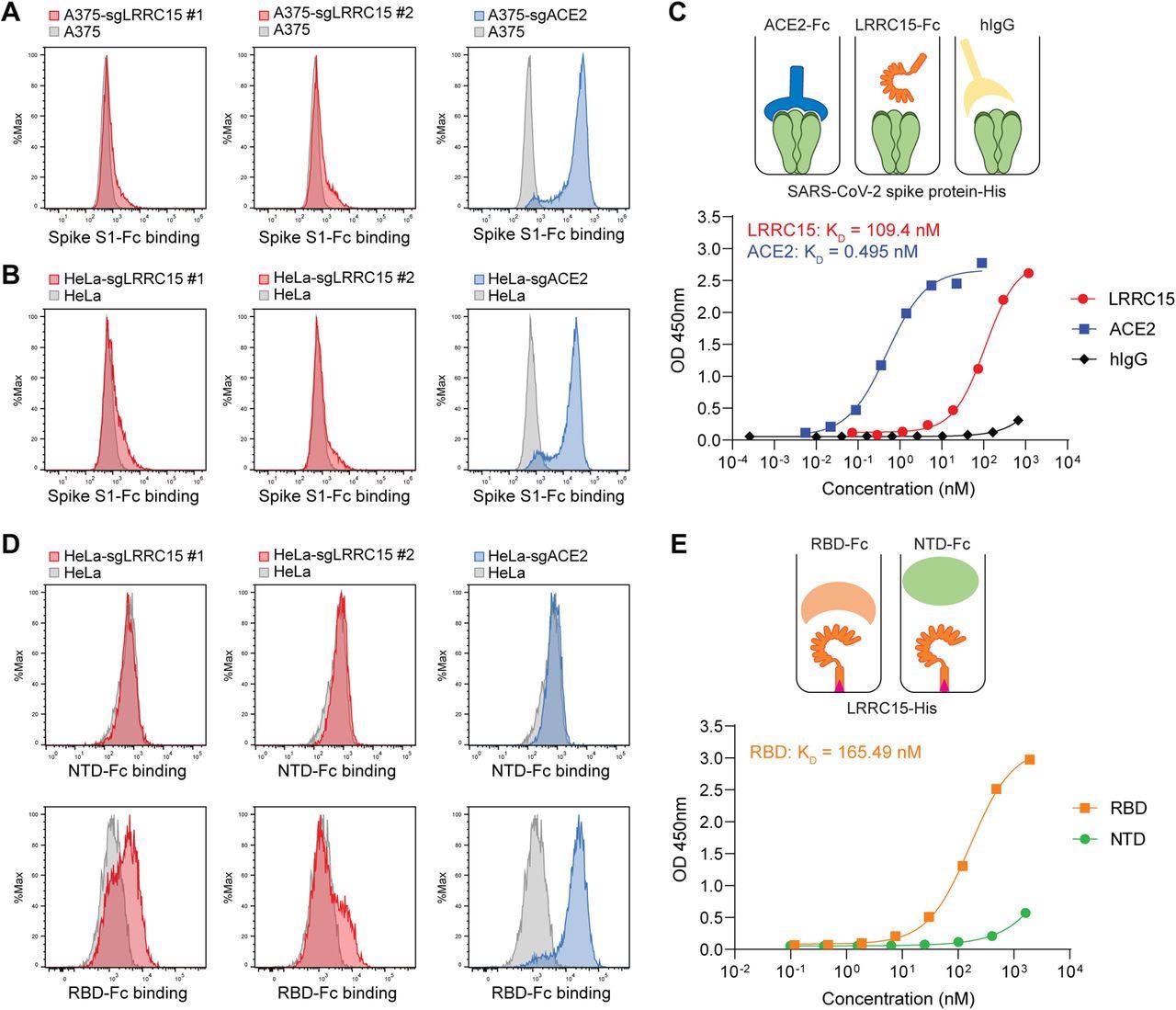 LRRC15 binds with SARS-CoV-2 spike protein at the receptor-binding domain (A) A375 cells were transduced with indicated activating sgRNAs and incubated with SARS-CoV-2 spike S1-Fc fusion protein. Protein binding was measured by flow cytometry. (B) HeLa cells were transduced with indicated activating sgRNAs and incubated with SARS-CoV-2 spike S1-Fc fusion protein. Protein binding was measured by flow cytometry. (C) Dose-dependent binding of SARS-CoV-2 spike protein (Wuhan-Hu-1) to both ACE2 and LRRC15 with a Fc tag was determined by ELISA. Human IgG1 was included as a negative control. Dots indicate means of duplicates. (D) HeLa cells were transduced with indicated activating sgRNAs and incubated with SARS-CoV-2 spike NTD-Fc or RBD-Fc fusion protein. Protein binding was measured by flow cytometry. (E) The binding of the SARS-CoV-2 RBD and NTD to LRRC15 was measured by ELISA.
LRRC15 binds with SARS-CoV-2 spike protein at the receptor-binding domain (A) A375 cells were transduced with indicated activating sgRNAs and incubated with SARS-CoV-2 spike S1-Fc fusion protein. Protein binding was measured by flow cytometry. (B) HeLa cells were transduced with indicated activating sgRNAs and incubated with SARS-CoV-2 spike S1-Fc fusion protein. Protein binding was measured by flow cytometry. (C) Dose-dependent binding of SARS-CoV-2 spike protein (Wuhan-Hu-1) to both ACE2 and LRRC15 with a Fc tag was determined by ELISA. Human IgG1 was included as a negative control. Dots indicate means of duplicates. (D) HeLa cells were transduced with indicated activating sgRNAs and incubated with SARS-CoV-2 spike NTD-Fc or RBD-Fc fusion protein. Protein binding was measured by flow cytometry. (E) The binding of the SARS-CoV-2 RBD and NTD to LRRC15 was measured by ELISA.
A notable observation was that besides the same cells, LRRC15 also inhibited the viral entry in neighboring cells in trans.
By interacting with the spike protein itself, the inhibition of viral entry by LRRC15 demonstrated a direct effector. In many cases, the LRR domain has been linked with sensing the pathogen-associated molecular patterns (PAMPs).
One notable feature of LRR domain proteins is that they are highly conserved (even in plants). This provides superior protection against pathogens.
It is possible that human beings could develop a pattern recognition protein for certain types of coronaviruses, given the role of LRR domains in pattern recognition.
The inhibitory effect of LRRC15 against coronaviruses signifies an arms race between humans and coronaviruses. Furthermore, at the time of preparation of the manuscript of the current research, LRRC15 was identified as a SARS-CoV-2 spike binding factor in two other studies, proving the robust and detectable nature of the LRRC15-spike interaction.
Many interferon-stimulated genes (ISGs), such as LY6E, CH25H, and IFITMs, have been shown to hinder the entry of coronaviruses by interfering with spike protein-mediated membrane fusion.
Alternatively, the ISGs could also interfere with endosome-mediated processes. However, the mechanism of action of ISGs and LRRC15 is quite different.
LRRC15 is not induced by interferons and the inhibiting action is directly mediated by interaction with the spike, which resembles PAMP receptors.
Scientists stated that LRRC15 could act as an antiviral factor via different modes as it redistributes adenovirus receptors, thereby, affecting the delivery of adenovirus to cells.
Future research will need to determine whether LRRC15 requires intracellular signaling (through the cytosolic domain) or other cellular proteins for the inhibition.
Researchers stated that the trans-inhibition of viral was notable. They co-cultured ACE2+LRRC15- cells and ACE2-LRRC15+ cells and observed a significant suppression of viral infection. This indicated that the antiviral effect of LRRC15 can be broad in a physiological context. In the human lung, during SARS-CoV-2 infection, LRRC15 could function as an entry inhibitor and act as a viral control.
Conclusion
Attachment factors and co-receptors facilitating SARS-CoV-2 entry have been extensively studied, but the cellular entry factors inhibiting viral entry are largely unknown.
The current study showed LRRC15 (by directly interacting with the spike protein) to be a receptor for the SARS-CoV-2 spike and represent a pattern-recognition-like inhibition of viral entry.
Therefore, the study provided novel insights for therapeutic development and a better understanding of COVID-19.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Song, J. et al. (2021) LRRC15 is an inhibitory receptor blocking SARS-CoV-2 spike-mediated entry in trans. bioRxiv 2021.11.23.469714; doi: https://doi.org/10.1101/2021.11.23.469714, https://www.biorxiv.org/content/10.1101/2021.11.23.469714v1
- Peer reviewed and published scientific report.
Song, Jaewon, Ryan D. Chow, Mario A. Peña-Hernández, Li Zhang, Skylar A. Loeb, Eui-Young So, Olin D. Liang, et al. 2022. “LRRC15 Inhibits SARS-CoV-2 Cellular Entry in Trans.” Edited by Tom Gallagher. PLOS Biology 20 (10): e3001805. https://doi.org/10.1371/journal.pbio.3001805. https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.3001805.