The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is the virus responsible for the coronavirus disease 2019 (COVID-19), primarily targets respiratory organs, as it reproduces in, and subsequently damages and kills, these epithelial cells. This leads to widespread inflammation and immune dysfunction, including issues such as cytokine storms.
In addition to these adverse effects of COVID-19, many patients will also report neurological symptoms such as memory loss, mental health issues, as well as cognitive and psychiatric disorders. These are most commonly reported in patients suffering from 'long COVID,' where the symptoms persist long after the initial infection.
 Study: Neurotoxic Amyloidogenic Peptides Identified in the Proteome of SARS-COV2: Potential Implications for Neurological Symptoms in COVID-19. Image Credit: Kateryna Kon / Shutterstock.com
Study: Neurotoxic Amyloidogenic Peptides Identified in the Proteome of SARS-COV2: Potential Implications for Neurological Symptoms in COVID-19. Image Credit: Kateryna Kon / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
In a recent study published on the preprint server bioRxiv*, researchers from La Trobe University investigate potentially amyloidogenic peptide fragments that could be neurotoxic in an effort to explain some of the neurological symptoms associated with COVID-19 and long-COVID.
About the study
In the current study, the researchers used two different algorithms to predict peptide sequences that showed a tendency to form beta-rich amyloid assemblies. TANGO is an algorithm used to predict aggregation nucleation regions in unfolding polypeptide chains, assuming that the aggregating regions are within the hydrophobic core, while ZIPPER predicts hexapeptides within larger polypeptide sequences.
When the ZIPPER tool was applied to open reading frame 6 (ORF-6), it showed more than ten choices of six-residue windows. These were narrowed down by also using the TANGO algorithm, which left two regions predicted to be highly aggregation-prone, of which include I14LLIIMR and D30YIINLIIKNL.
ILLIIM was chosen as the first candidate, as it closely resembles a sequence from Hen Egg White Lysozyme, which has also been seen to be highly amyloidogenic. TANGO plots for ORF-10 shows that the main aggregation-prone sequence is residues F11TIYSLLLC; however, this was not confirmed by ZIPPER.
Next, the researchers chose the octapeptide R24NYIAQVD due to its zwitterionic residue pair R-D, which appears to strongly enhance interpeptide association. A hexapeptide within RNYIAQVD was also predicted to be highly amyloidogenic by ZIPPER. The scientists then decided to synthesize and investigate RNYIAQVD and ILLIIM.
Study findings
Atomic force microscopy (AFM) and transmission electron microscopy (TEM) imaging shows both peptides can assemble into needle-like crystalline assemblies in as little as two hours at significant concentrations. Both peptides tend to stack on top of each other in order to form multilaminar nonfibrillar structures, which occurs more often in RNY1AQVD than in ILLIIM.
ILLIM varies between 4-9 nanometers (nm) tall, while RNY1AQVD is an average height of 5.5 nm. ILLIIM is also very wide at around 2-3 microns in length.
Statistical analysis of fibril widths and contour lengths revealed that both peptides show a heterogeneous distribution of fibril widths and a biphasic distribution of lengths, with two broad sub-populations centered around 1 and 3 micrometers (µm). The amyloid nature of the two assemblies was further confirmed by wide angle X-ray scattering (WAXs) spectra, with both peptides possessing a number of strongly diffracting Bragg peaks, including a characteristic peak at 1.38A-1, which corresponds to a d-spacing of A that is indicative of amyloid assembly from extended beta-sheets.
The researchers argue that their hypothesis that these two viral transcript fragments are toxic to human neurons is supported by their findings. This neurotoxic potential is corroborated by previous reports on the neuroinvasive capabilities of SARS-CoV-2, coupled with the similarities of the symptoms to Alzheimer's disease and the previous detection of any amyloid assemblies driven by other viruses.
Cytotoxic assays of the two peptides against a human-derived neuroblastoma cell line (SH-SY5Y) revealed that both assemblies were highly toxic after 48-hour incubation with the target peptide. Concentrations as low as 0.05/0.04 millimolar (mM) were seen to kill over 50% of the cell lines, which are often used as a model cell line for studying neurodegenerative disorders.
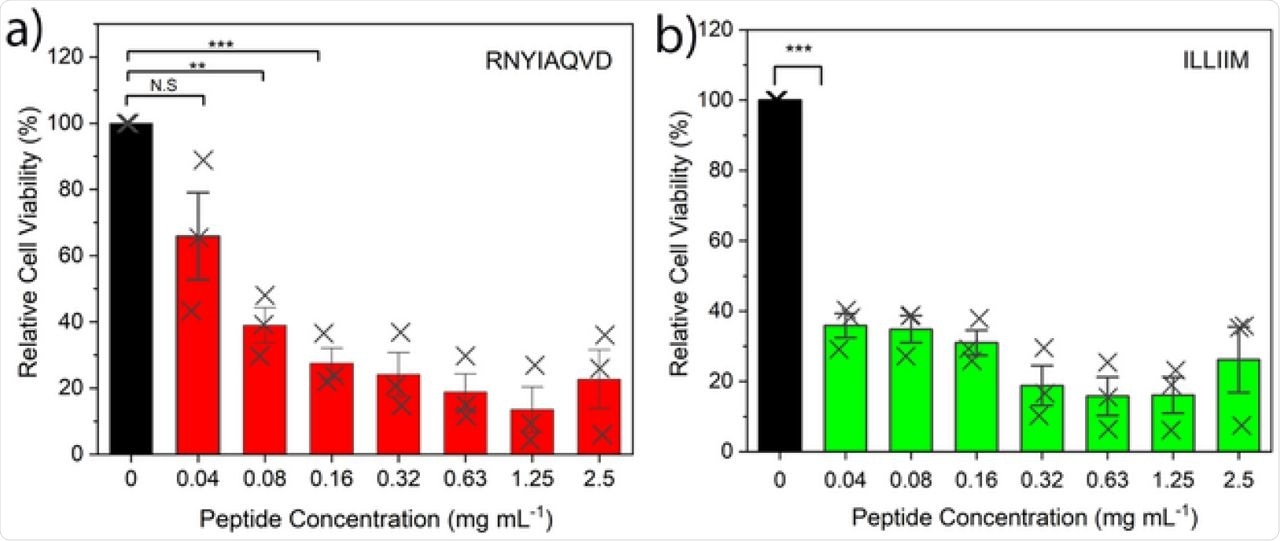 Cytotoxicity assays of RNYIAQVD and ILLIIM assemblies over a range of concentrations. Error bars represent 1 Standard Deviation. Statistical analysis performed by one way ANOVA with Tukey comparison. N.S. = No statistical significance, ** = p < 0.0005, *** = p < 0.0001.
Cytotoxicity assays of RNYIAQVD and ILLIIM assemblies over a range of concentrations. Error bars represent 1 Standard Deviation. Statistical analysis performed by one way ANOVA with Tukey comparison. N.S. = No statistical significance, ** = p < 0.0005, *** = p < 0.0001.
Conclusions
The scientists urge further investigations into the presence of amyloid aggregates from SARS-CoV-2 in the central nervous system (CNS) of COVID-19 patients and suggest that these may be responsible for some of the neurological symptoms observed in these patients. This would explain some of the symptoms seen in 'long COVID,' in which patients suffer a wide variety of symptoms, including long-term anosmia, fatigue, depression, anxiety tinnitus, and earaches.
This information could help to inform healthcare workers and researchers, as well as possibly lead to a treatment or preventative treatment to help prevent these symptoms from developing in others.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Islam, S., Charnley, M., Bindra, G., et al. (2021). Neurotoxic Amyloidogenic Peptides Identified in the Proteome of SARS-COV2: Potential Implications for Neurological Symptoms in COVID-19. bioRxiv. doi:10.1101/2021.11.24.469537. https://www.biorxiv.org/content/10.1101/2021.11.24.469537v1
- Peer reviewed and published scientific report.
Charnley, Mirren, Saba Islam, Guneet K. Bindra, Jeremy Engwirda, Julian Ratcliffe, Jiangtao Zhou, Raffaele Mezzenga, et al. 2022. “Neurotoxic Amyloidogenic Peptides in the Proteome of SARS-COV2: Potential Implications for Neurological Symptoms in COVID-19.” Nature Communications 13 (1). https://doi.org/10.1038/s41467-022-30932-1. https://www.nature.com/articles/s41467-022-30932-1.