As one of the twenty-first century's most significant global health issues, the ongoing severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has indeed altered our lives. Unlike earlier pandemics, however, we now have high-throughput sequencing capabilities to investigate the genome composition of SARS-CoV-2. In addition, we can track and define the viral genome evolution for near real-time surveillance as labs around the world sequence isolates from infected individuals.
VAPiD, Prokka, InterProScan, and other viral genome annotation technologies, for example, attempt to offer autonomous annotation of genes and proteins.
Special releases of some of these tools have been made to aid in the annotation of SARS-CoV-2 genomes. However, many of these methods are designed for more general uses, do not provide sufficient accuracy with "off the shelf" SARS-CoV-2 use, and have not yet been deployed at scale as the amount of SARS-CoV-2 sequence data grows.
Several SARS-CoV-2 genome variants have also been developed, including the D614G variant, which first appeared earlier in the pandemic, and the more recent B.1.1.7 (Alpha) or B.1.617.2 (Delta) variants, which account for the bulk of new cases in the United States and around the world. Unfortunately, the mutations that define these variants can make comprehensive genome annotation difficult, and the SARS-CoV-2 transcriptional slippage site can make things even more difficult.
In a new research paper, a group of scientists from various institutions offers a semi-supervised custom pipeline for annotating all SARS-CoV-2 genes, proteins, and functional domains. These sequences serve as the molecular targets for better diagnostics, antivirals, and vaccinations. This semi-supervised technique was used to analyze 66,905 SARS-CoV-2 genomes from the NCBI GenBank and GISAID databases. The IBM Functional Genomics Platform, a technology made publicly available to the COVID-19 worldwide research community, revealed almost 13 million unique molecular sequences and linkages as a result of this method.
This study is available in the journal Viruses.
 Study: Semi-Supervised Pipeline for Autonomous Annotation of SARS-CoV-2 Genomes. Image Credit: NIAID
Study: Semi-Supervised Pipeline for Autonomous Annotation of SARS-CoV-2 Genomes. Image Credit: NIAID
The study
A COVID-19 genome annotation pipeline must reliably identify all known molecular targets inside a genome to be clinically and biologically useful. The SARS-CoV-2 proteome consists of thirteen protein products, each with a corresponding gene sequence in each genome. SARS-CoV-2 proteins are classified as structural or non-structural, but all are essential for the virus's life cycle, which involves host cell invasion, replication, and transmission.
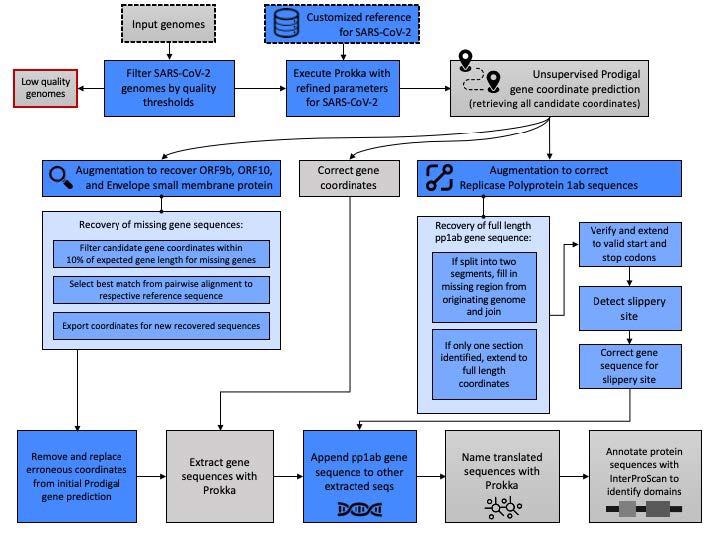 Pipeline schematic for semi-supervised identification of molecular targets. The data and analytic steps comprising our SARS-CoV-2 annotation pipeline described in Methods Sections 2.1–2.3 are indicated. Input data are indicated with a dashed line and discarded data, e.g., genomes below quality thresholds, are indicated with a red line. Blue boxes indicate steps where our augmentation of base Prokka and Prodigal yield methodological advancements specific to the requirements of SARS-CoV-2 genomes.
Pipeline schematic for semi-supervised identification of molecular targets. The data and analytic steps comprising our SARS-CoV-2 annotation pipeline described in Methods Sections 2.1–2.3 are indicated. Input data are indicated with a dashed line and discarded data, e.g., genomes below quality thresholds, are indicated with a red line. Blue boxes indicate steps where our augmentation of base Prokka and Prodigal yield methodological advancements specific to the requirements of SARS-CoV-2 genomes.
Across all genomes over the aforementioned quality levels, the authors achieved an average per protein identification accuracy of 98.5 ± 2.9% using their gene and protein annotation method. The researchers were able to attain entire or near-complete protein set membership for all genomes based on the number of observations per protein. Because each protein is a translated gene sequence, the same level of gene identification precision is attained.
Furthermore, for accurate genome annotation, not only must the entire set of designated genes and proteins be identified, but the created sequences must also be based on biological reality. Given the relatively recent development of SARS-CoV-2 and documented lower mutation rate than other RNA viruses, in silico projected sequences should not be trimmed compared to the length of known references, and mutational density must be low.
The authors were able to identify full-length protein products that, on a per protein basis, match the expected lengths of known reference sequences with an average observed/expected protein length value of 99.1% using the semi-supervised gene and protein annotation method. Furthermore, by using a two-sample Kolmogorov–Smirnov test, the distributions of the projected and anticipated protein sequence lengths are found to be statistically similar. They are 8.75-fold more similar than those predicted from genomes that do not meet the quality requirements, i.e., poor quality genomes.
The method utilized in this study was able to identify 6.4-fold more protein products compared to base Prokka and was able to generate full-length pp1ab products with high sequence identity to known UniProt references.
The authors compared their pipeline to VAPiD, which generated a specific release for annotating SARS-CoV-2 genomic data, and Prokka, a prokaryotic genome annotation tool for bacteria and viruses, in terms of pipeline accuracy. The obtained protein annotations in terms of set membership, as well as observed protein sequence length versus reference protein sequence length, were examined using the same collection of genomes.
Both VAPiD and the author's technique obtained good accuracy in truncated proteins, but the pipeline elicited 1.8-fold more proteins in the highest accuracy category and 1.8-fold more protein annotations overall. The open reading frame (ORF) 9b and Protein 3a were consistently absent from VAPiD annotations. Prokka, on the other hand, yielded no full-length pp1ab protein sequences and produced a large number of missing or truncated proteins, particularly for Envelope small membrane protein, ORF9b, and ORF10, among other proteins.
Implications
This approach can be used to monitor and track developing protein variations across hosts swiftly and sampling niches, such as aerosol, wastewater, and surfaces, to inform disease understanding, vaccine specificity, and host protein binding affinity as vaccination rates grow, and the pandemic persists.
Furthermore, future studies will refine the protein sequences and important domains to increase the understanding of interactions with host proteins, antivirals, or diagnostics by utilizing a structural model to corroborate the in silico predicted sequences. Overall, the data obtained as part of this project give a comprehensive database of protein and domain variants observed around the world, which will aid researchers in their efforts to understand and contain the COVID-19 pandemic.
Journal reference:
- Semi-Supervised Pipeline for Autonomous Annotation of SARS-CoV-2 Genomes, Kristen L. Beck, Edward Seabolt, Akshay Agarwal, Gowri Nayar, Simone Bianco, Harsha Krishnareddy, Timothy A. Ngo, Mark Kunitomi, Vandana Mukherjee and James H. Kaufman, MDPI, 2021.12.03, https://doi.org/10.3390/v13122426, https://www.mdpi.com/1999-4915/13/12/2426