As the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continues to mutate and spread across the world, various types of surveillance are necessary to monitor outbreaks and the emergence of new variants. In particular, these strategies are key to identifying SARS-CoV-2 variants of concern (VOCs) that are resistant to neutralization by the polyclonal antibodies elicited by natural infection or by vaccination.

Study: An Antibody-Escape Calculator for Mutations to The Sars-Cov-2 Receptor-Binding Domain. Image Credit: ETAJOE / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
When a pandemic is progressing, time is of the essence. SARS-CoV-2 has already mutated many times, with clusters of mutations occurring at key antigens, notably the spike glycoprotein. These pose challenges to pre-existing immunity in vaccinated or recovered individuals.
The spike protein is responsible for viral binding to the angiotensin-converting enzyme 2 (ACE2) receptor and viral entry into the host cell via endocytosis that is mediated by virus-cell membrane fusion. Mutations in the spike protein, especially at the receptor-binding domain (RBD), reduce viral neutralization by antibodies, as shown by the Beta and Delta VOCs.
Much time and effort are required to understand the extent to which immunity is compromised by these mutations using neutralization assays. This has caused delays in the characterization of most of these new variants, beyond the speed with which they are identified by genomic sequencing.
Deep mutational scanning experiments are one way to avoid this lag, as these can be designed to assess mutational impacts on antibody binding or neutralization for new mutations. The goal of these experiments is to examine every possible change at each residue in the important functional regions of the spike antigen to determine the effect it could have on the antigenic properties when encountering a monoclonal-specific antibody.
This approach is difficult to adopt in the case of the SARS-CoV-2 spike antigens, as VOCs typically show multiple mutations. Furthermore, the effects of these mutations are difficult to assess in combination, even with this relatively high-throughput experimental method.
About the study
In response, the researchers in this study collected previously published deep mutational scanning data for a number of antibodies to determine how mutations in the RBD of the spike effect neutralization.
This escape calculator tool allows investigators to understand how random combinations of antigens affect the antigenicity of the spike RBD, in visual as well as quantitative terms. The calculator uses simple transformations of experimental data to map combined escape mutational effects without relying on complicated calculations.
By taking the sum or average of all mutational effects due to single-residue mutations at the RBD, in terms of monoclonal antibody binding by the antigen, the calculator produces a summary of escape from antibodies at each site.
Study findings
Using site-specific escape maps, the researchers found that these monoclonal antibody escape maps showed which mutations are correlated with each other and which are independent in their effects. For instance, three RBD-neutralizing monoclonal antibodies of LY-CoV016 (etesevimab), LY-CoV555 (bamlanivimab), and REGN10987 (imdevimab) have been affected by mutations at each RBD site.
Earlier studies have described these changes in binding for each site. These antibodies bind distinct sites, or epitopes, on the RBD including LY-CoV016 to the class 1 epitope, LY-CoV555 the class 2 epitope, and REGN10987 the class 3 epitope. As a result, different sets of mutations are required to produce escape from neutralization by each of these antibodies.
Thus, mutations at residue 417 cause LY-CoV016 escape; site 484 for LYCoV555, and sites 444–446 for REGN10987. This means that a cocktail with equipotent concentrations of all three could be rendered ineffective by looking at the escape maps for each of these. This is the escape map for the antibody mix.
This polyclonal escape map will show the highest peaks at the sites where mutational escape to the individual antibodies is greatest. This can now be transformed to understand what would happen if one of these antibodies is rendered non-neutralizing, simply by inserting a mutation at its epitope. The peaks at this site will vanish, as will the peaks at other sites that are also bound by the same antibody.
This allows the researchers to visualize the correlations between the epitope targeted by the mutation and other sites that recognize antibodies that bind to this epitope as part of its target binding site. Other escape epitopes remain intact since they are bound by other antibodies.
If a second antibody is also eliminated by mutations at its epitope, the two correlated peaks on the escape map will also disappear. Therefore, different possibilities can be explored with this tool.
The authors of this study encourage other researchers to try different scenarios to understand how this tool works. They have collected deep mutational scanning data for 33 human antibodies to produce an escape calculator that will help understand realistic possibilities for mutational escape from individual antibodies or from a polyclonal mix that resembles human sera.
The 33 antibodies used here are from multiple patients infected during the first year of the coronavirus disease 2019 (COVID-19) pandemic. While assuming that an equipotent mix of these would be equivalent to the neutralizing activity of human sera for experimental purposes, the researchers point out that this is unlikely to be the case.
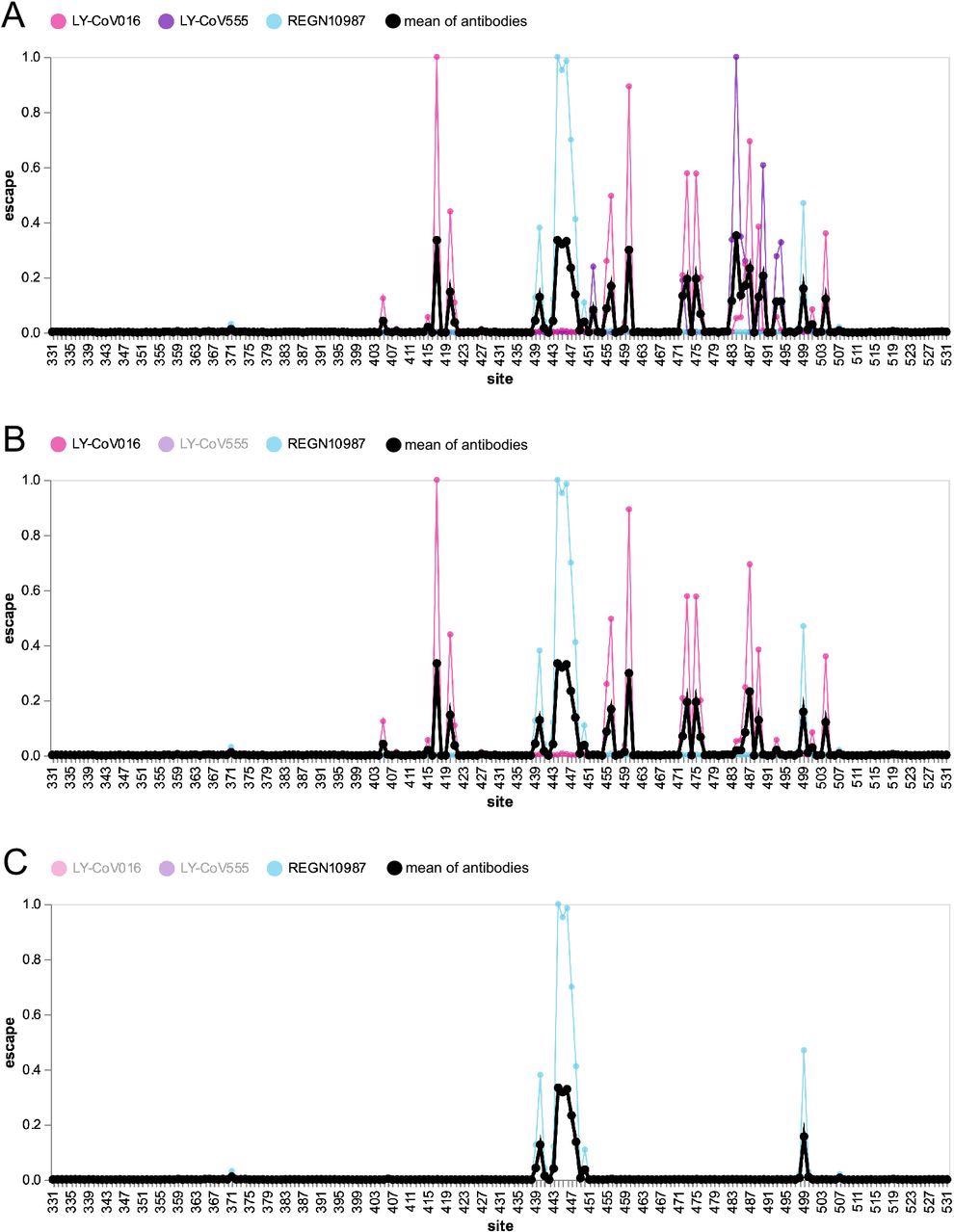 Escape map for a hypothetical polyclonal mix consisting of an equipotent mixture of three monoclonal antibodies targeting distinct epitopes on the SARS-CoV-2 RBD. (A) Experimentally measured escape maps for three antibodies, and the mean of these maps (thick black line). Each point on the x-axis represents a site in the RBD, and the y-axis represents the total measured escape by all mutations at that site scaled so the maximum for each antibody is one. (B) Escape map if the contribution of antibody LY-CoV555 is ablated.(C) Escape map if the contributions of antibodies LY-CoV555 and LY-CoV016 are ablated. An interactive version of this figure is at https://jbloomlab.github.io/SARS2_RBD_Ab_escape_maps/mini-example-escape-calc/.
Escape map for a hypothetical polyclonal mix consisting of an equipotent mixture of three monoclonal antibodies targeting distinct epitopes on the SARS-CoV-2 RBD. (A) Experimentally measured escape maps for three antibodies, and the mean of these maps (thick black line). Each point on the x-axis represents a site in the RBD, and the y-axis represents the total measured escape by all mutations at that site scaled so the maximum for each antibody is one. (B) Escape map if the contribution of antibody LY-CoV555 is ablated.(C) Escape map if the contributions of antibodies LY-CoV555 and LY-CoV016 are ablated. An interactive version of this figure is at https://jbloomlab.github.io/SARS2_RBD_Ab_escape_maps/mini-example-escape-calc/.
The escape map shows the interesting fact that the escape peak at 484 is the largest of all since antibodies to this epitope were most common in the immune humoral response during this period. Many smaller peaks were also observed at other sites, as antibodies tend to bind to somewhat unpredictable antigenic sites.
By introducing mutations at different sites and reducing the peaks in relation to the strength of antibody binding at each site, it would be possible to understand what the escape map would look like for such a mix and predict how compound mutations would affect neutralization.
In the Beta VOC, for instance, three RBD sites (417, 484, and 501) are altered. The resulting escape map for this VOC, therefore, exhibits loss of binding from the mutated sites and others that target epitopes, including these sites. Other antibodies targeting sites 444–446, for example, continue to show neutralizing activity because they do not bind to the above sites.
The escape map fits actual neutralization data from experimental neutralization assays using human sera against SARS-CoV-2 variants and allows quantitative scoring of residual polyclonal antibody binding for each set of mutations.
“The simple and intuitive approach used by the calculator seems to accurately reflect the dominant features of polyclonal antibody escape in the RBD.”
With respect to the new Omicron variant, the escape calculator indicates a wide-ranging change in the binding epitopes due to its 15 mutations in the RBD. The binding score provided by the calculator is very low compared to all other variants so far.
This suggests that the Omicron variant will resist neutralization by existing antibodies. Furthermore, this variant was provided a score similar to that of an engineered spike protein with multiple mutations intended to provide a picture of the maximum possible antibody escape mutational profile at 20-80-fold reduced neutralization by sera from infected-recovered or vaccinated individuals.
The greatest loss of antibody binding is at sites 484, 446, and 417, whereas other mutations at sites like 346, 378, 444, and 504 could eliminate whatever is left.
Implications
The escape calculator presented here, which is based on experimental data for 33 monoclonal antibodies, aims to predict the antigenic effects of sets of mutations to the SARS-CoV-2 RBD. It was designed on the basis of aggregating this data to identify the most vital antigenic sites in the RBD and distinguish correlated and independent mutations.
This interactive tool provides a visual representation that allows easy understanding of the effects of single or combined mutations by clicking on different sites.
“This interactivity provides an intuitive understanding of the antigenic structure of the RBD, and shows how the calculator is performing simple transformations directly on experimental data.”
Despite the limitations of this early model, it was able to produce scores of binding activity that bore an uncanny resemblance to actual experimental neutralization activity. Moreover, the shift in antigenic sites occurs in some manner similar to the underlying design of the escape calculator, such as the presence of clustered mutations that produce additive rather than redundant escape effects in VOCs.
This may be due to the fact that the escape calculator is based on RBD mutations as compared to antibodies to the ancestral SARS-CoV-2 variant. The RBD is the dominant target of neutralizing antibodies, with a common antibody response observed against the early viral variants worldwide, thus providing common ground for experimental data and this calculator.
The scientists point out that the reference sequence to determine mutations is shifting month by month as the pandemic progresses, with respect to antibody escape. This will make it necessary to classify serum antibody data used in the calculator by RBD variant. To study the escape map for each such case, only that antibody data that is relevant to the sera being studied will have to be gathered.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources