The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is the virus responsible for the coronavirus disease 2019 (COVID-19), has been circulating around the world ever since it emerged towards the end of 2019. The COVID-19 pandemic has caused unforeseeable distress, illness, and death around the world, which has led to the rapid development of vaccines in an attempt to contain the pandemic.
Study: Production and Secretion of Functional Full-Length SARS-Cov-2 Spike Protein In Chlamydomonas Reinhardtii. Image Credit: Achiichii / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
The spike protein, which is studded over the surface of SARS-CoV-2, is targeted by 55 neutralizing antibodies and is the primary antigenic basis of all currently available vaccines against this virus. The SARS-CoV-2 spike protein is a glycoprotein that consists of over 1,200 residues, with 22 sugars attached through N-linkages. This makes it difficult to produce the spike protein by recombination through any other platform than mammalian and insect cells, thereby rendering it essential to use complex culture media.
Glycosylation is among the most commonly observed post-translational modification in the synthesis of biopharmaceuticals. This is a vital component of functional protein synthesis, as it is required for the proper folding of proteins and their stability.
Among the current COVID-19 vaccines, the messenger ribonucleic acid (mRNA) and adenovirus vector vaccines have garnered the greatest interest. However, both vaccine platforms have been associated with serious, albeit rare, adverse effects, such as myocarditis and blood clots. Moreover, the development of these vaccines requires sophisticated technology, making them unsuitable for less developed countries.
About the study
In the current paper, the researchers examined the potential for the production of full-length spike protein, rather than only the receptor-binding domain (RBD), since many neutralizing antibodies bind outside the RBD.
Herein, the researchers attempted to produce a simple platform for the production of the full-length spike and possibly other therapeutic glycoproteins, thus facilitating vaccine development in poorer regions. They chose microalgae, which grow rapidly and profusely in very simple culture media, with no toxic byproducts.
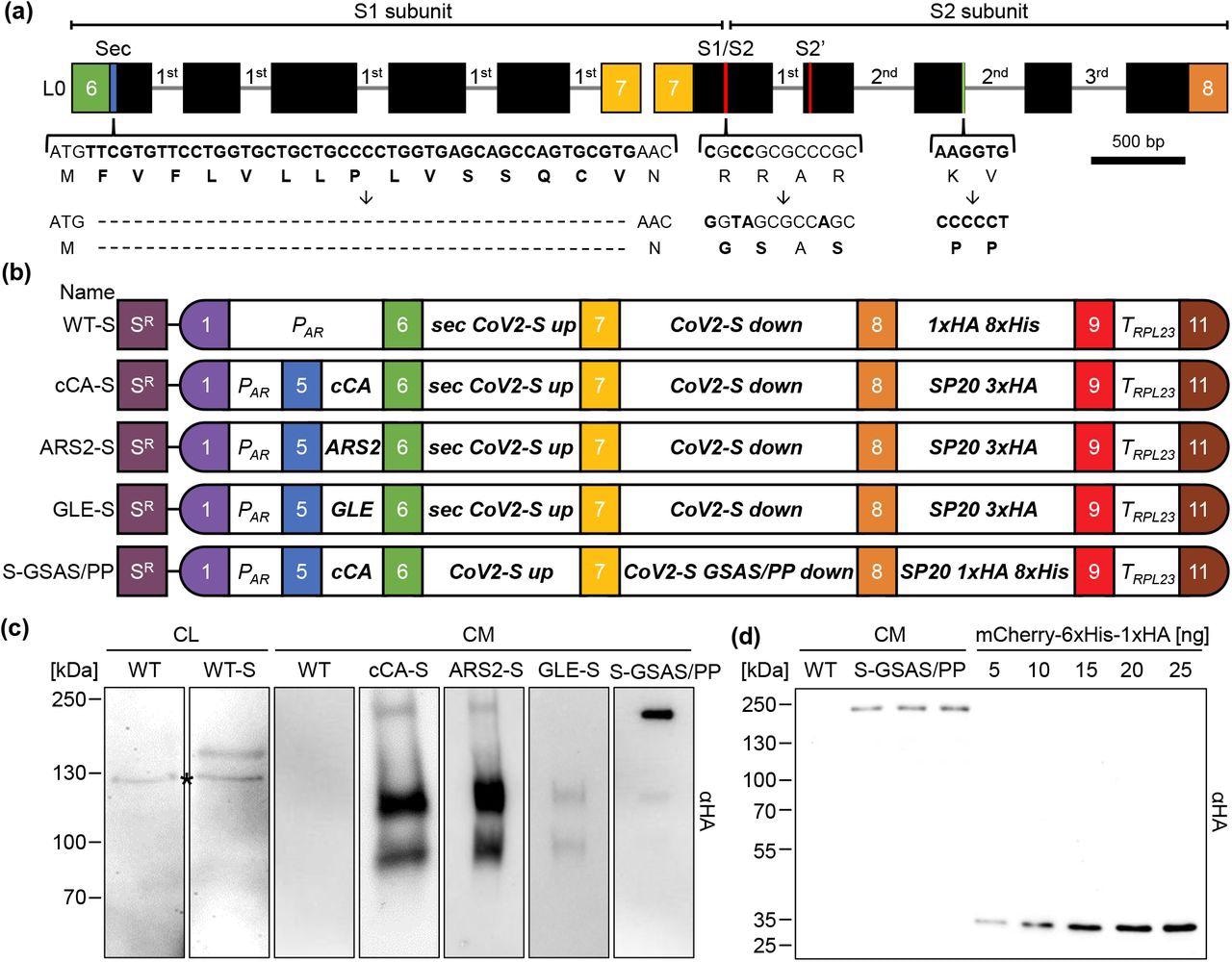 Design of constructs for the production and secretion of the SARS CoV-2 spike protein in Chlamydomonas. (a) Schematic representation of the two-level 0 parts encoding the spike protein. The codon adapted coding region is depicted by black boxes and incorporated RBSC2 introns by thin lines. The braces highlight regions modified by site-directed mutagenesis and the resulting amino acid modifications. Colored boxes represent fusion sites according to the MoClo syntax. (b) Level 2 MoClo devices consist of a level 1 module containing the aadA resistance marker under the control of the PSAD promoter and terminator (SR), and level 1 transcriptional units for the expression of the spike protein. The latter contains the HSP70A-RBCS2 promoter (PAR), sequences encoding various secretion signals (cCA, ARS, GLE, sec), the two parts of the spike protein (CoV2-S up and down), a glycomodule of 20 serine/proline repeats (SP20) fused to the HA epitope and/or an octa-his tag, and the RPL23 terminator (TRPL23). (c) Production and secretion of the spike protein in transformants generated with the five level 2 devices listed in (b). Shown are representative immunoblots detecting HA-tagged proteins in cell lysates (CL) (corresponding to 2 μg chlorophyll) and 1.7 mL of culture medium (CM) after TCA precipitation. The asterisk indicates a cross-reaction of the HA antibody. (d) Three independent preparations of spike protein secreted by a S-GSAS/PP transformant into 1.7 mL culture medium (CM) and precipitated with TCA, and increasing amounts of purified recombinant mCherry carrying a C-terminal HA tag were analyzed by immunoblotting using an HA antibody.
Design of constructs for the production and secretion of the SARS CoV-2 spike protein in Chlamydomonas. (a) Schematic representation of the two-level 0 parts encoding the spike protein. The codon adapted coding region is depicted by black boxes and incorporated RBSC2 introns by thin lines. The braces highlight regions modified by site-directed mutagenesis and the resulting amino acid modifications. Colored boxes represent fusion sites according to the MoClo syntax. (b) Level 2 MoClo devices consist of a level 1 module containing the aadA resistance marker under the control of the PSAD promoter and terminator (SR), and level 1 transcriptional units for the expression of the spike protein. The latter contains the HSP70A-RBCS2 promoter (PAR), sequences encoding various secretion signals (cCA, ARS, GLE, sec), the two parts of the spike protein (CoV2-S up and down), a glycomodule of 20 serine/proline repeats (SP20) fused to the HA epitope and/or an octa-his tag, and the RPL23 terminator (TRPL23). (c) Production and secretion of the spike protein in transformants generated with the five level 2 devices listed in (b). Shown are representative immunoblots detecting HA-tagged proteins in cell lysates (CL) (corresponding to 2 μg chlorophyll) and 1.7 mL of culture medium (CM) after TCA precipitation. The asterisk indicates a cross-reaction of the HA antibody. (d) Three independent preparations of spike protein secreted by a S-GSAS/PP transformant into 1.7 mL culture medium (CM) and precipitated with TCA, and increasing amounts of purified recombinant mCherry carrying a C-terminal HA tag were analyzed by immunoblotting using an HA antibody.
Study findings
The experiment demonstrates the successful establishment of the unicellular green alga Chlamydomonas reinhardtii as a platform for the production and secretion of the full-length SARS-CoV-2 spike without the 19-C terminal portion, which acts as a membrane anchor. The spike protein’s native signal peptide is not sufficient to chaperone the spike protein to the secretory pathway within the Chlamydomonas cell. The scientists, therefore, used that of the algal cell itself.
During its trafficking through the secretory pathway, the spike protein was found to undergo post-translational cleavage at the furin cleavage site between the two subunits. This mirrored the protein processing within mammalian cells, which indicates that the endoplasmic reticulum and Golgi system in this organism also contain a furin-like protease.
In addition, an O-glycan-linked site found near the furin cleavage site is thought to regulate this cleavage. This suggests that the Chlamydomonas cell also performs O-glycosylation, thus accounting for the efficient cleavage.
The scientists identified a 20 serine-proline repeats (SP20) segment that appears to be essential for proper spike secretion from Chlamydomonas. This corroborates earlier reports of protein secretion in this organism driven by this module.
Affinity purification of the spike protein by attaching a hemagglutinin or octahistidine tag at the C-terminal end did not succeed. This may be because the tag was removed by proteolysis while the protein was passing through the secretory pathway or in the medium, the tag was improperly folded and subsequently caused steric hindrance, or because other secreted proteins disrupted this process.
These algae secrete the protein stabilized in its prefusion form, cleaved at the furin cleavage site during its synthesis.
When this cleavage site is eliminated, cleavage does not occur, making it non-functional. The spike protein was enriched from the culture medium, using ammonium sulfate precipitation, and was found to be functional, as shown by its binding with the recombinant angiotensin-converting enzyme 2 (ACE2), as well as on the receptor expressed on human cells.
This also shows that proper N-glycosylation has taken place, though it seems to proceed beyond the SP20 module. The glycan linkages are not exactly like those observed when the spike protein is produced in human cells, leaving room for further engineered proteins to reflect a more humanized form of the Chlamydomonas spike.
Implications
Chlamydomonas reinhardtii is a microalgal generally recognized as a safe (GRAS) organism that is simple and inexpensive to produce at a large scale, as it requires only simple culture media. This makes it suitable for the production of recombinant full-length spike protein production, as well as other biopharmaceuticals in poor countries.
To date, mammalian and insect cells have been used as platforms for full-length spike production. The RBD alone has been produced within Chlamydomonas; however, this organism’s range has been expanded in the experiments conducted in the current study.
The method discussed here exploits the short generation time, easy transformation, and other appropriate strategies to facilitate rapid assembly and testing of spike glycoprotein constructs. Further, this approach should be easy to adapt for the production of spike variants present in the currently circulating viral strains of SARS-CoV-2 such as the Beta, Delta, and Omicron variants.
Further work must focus on efficient purification of the secreted spike protein and increased efficiency of N-linked glycosylation and protein folding, which is more difficult because of the large size of the protein with its abundant glycans. This will help secrete a more functional spike protein. Optimization of the culture conditions will help enhance the yield still further.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Kiefer, A., Niemeyer, J., Probst, A., et al. (2021). Production and Secretion of Functional Full-Length SARS-Cov-2 Spike Protein In Chlamydomonas Reinhardtii. bioRxiv. doi:10.1101/2021.12.13.472433. https://www.biorxiv.org/content/10.1101/2021.12.13.472433v1.
- Peer reviewed and published scientific report.
Kiefer, Anna Maria, Justus Niemeyer, Anna Probst, Gerhard Erkel, and Michael Schroda. 2022. “Production and Secretion of Functional SARS-CoV-2 Spike Protein in Chlamydomonas Reinhardtii.” Frontiers in Plant Science 13 (September). https://doi.org/10.3389/fpls.2022.988870. https://www.frontiersin.org/articles/10.3389/fpls.2022.988870.