The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) caused the emergence of the coronavirus pandemic (COVID-19) that has caused over five million deaths and caused hundreds of millions of infections. As a result, vaccine development efforts and monoclonal antibodies have been developed to treat and prevent the further spread of the virus and the progression of disease.
However, the emergence of new variants threatens the success of both these measures due to mutations in the viral spike that engages with neutralizing antibodies and the host cell receptor, the angiotensin-converting enzyme 2 (ACE2) receptor.
A new preprint research paper posted to the bioRxiv* server examines the nature and effects of the spike mutations and identifies a vulnerable point that could help develop broad-spectrum protection against circulating variants.
 Study: Emerging SARS-CoV-2 Variants of Concern: Spike Protein Mutational Analysis and Epitope for Broad Neutralization. Image Credit: NIAID
Study: Emerging SARS-CoV-2 Variants of Concern: Spike Protein Mutational Analysis and Epitope for Broad Neutralization. Image Credit: NIAID

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
While the virus spread worldwide, mutations began to be identified that were linked to increased transmissibility. The best-known was the D614G, in the spike glycoprotein, which led to its rise to dominance early in 2020. However, by November of the same year, the Alpha variant began to be widely identified and triggered a fresh spike of cases, first in the UK and then worldwide, to replace D614G as the globally dominant lineage.
For this reason, it was designated a variant of concern (VOC) of SARS-CoV-2. This was followed by a flurry of other VOCs, such as the Beta, Gamma, Epsilon, Kappa, and Delta. The last-named rapidly rose to dominance in the first quarter of 2021.
These variants were all found to be more transmissible than the wild-type variant, primarily due to their higher affinity for ACE2 due to the common N501Y mutation. Other commonly found mutations include E484K and K417T/N, both within the virus's receptor-binding domain (RBD) that impact ACE2 binding and escape from antibody-mediated neutralization. Other mutations in the N-terminal domain (NTD), within antigenic loop regions also enable antibody escape.
The current study reports on using an antibody called ab6 that neutralizes multiple variants and describes the epitope for this antibody in the RBD with cryogenic electron microscopy (cryoEM).
What Did the Study Show?
The antibody used here, VH ab6, has a higher affinity for the RBD in monomeric rather than bivalent form, an unusual antibody property. In addition, it has been shown to neutralize several RBD variants despite the presence of several mutations.
After confirming these properties in pseudovirus and live virus neutralization assays, they examined the ability of this antibody to bind to and neutralize Delta, Kappa, and other spike variants in circulation. The researchers found that it successfully binds to and neutralizes all the spike variants in pseudovirus assays, with lower neutralization potency for the Epsilon, Kappa, and Delta spikes.
Again, in live assays, Delta was less potently neutralized than the Alpha and Beta variants.
The crystal structure of ab6 was examined in complex with the wild-type spike. This showed binding in both up and down positions of the RBD, indicating a specialized mode of binding for this antibody, mostly via contacts between the beta-sheet ab6 scaffold that enfolds the RBD. This large area of interaction includes CDR2 and CDR3, but not CDR1.
The almost vertical level of interaction between the two surfaces means that only one heavy chain variable region (VH) can fit into the bivalent fusion molecule. This could be why the VH-Fc fusion construct acts as it does, relative to the VH ab6.
The area of binding includes L452, indicating that this accounts for reduced potency against all variants with the L452R mutation, namely, the Epsilon, Delta, and Kappa spikes. Conversely, this is the only mutation of all known variants that fall within the ab6 epitope, which doubtless explains the antibody's broad spectrum of neutralization.
Using globally uploaded sequencing data for SARS-CoV-2, it is clear that the ab6 epitope is highly conserved with only a single significant mutation, the L452R.
When the Epsilon spike-ab6 complex was examined by cryoEM, it was demonstrated that the antibody bound up and down RBDs just as with the wild-type spike. Further, it was shown that the 452R mutation causes extension of this position towards the scaffold, bringing this sidechain close to a hydrophobic region of ab6 around F58 and creating steric hindrance and electrostatic interference. This could account for the lower neutralization of spike variants with this mutation.
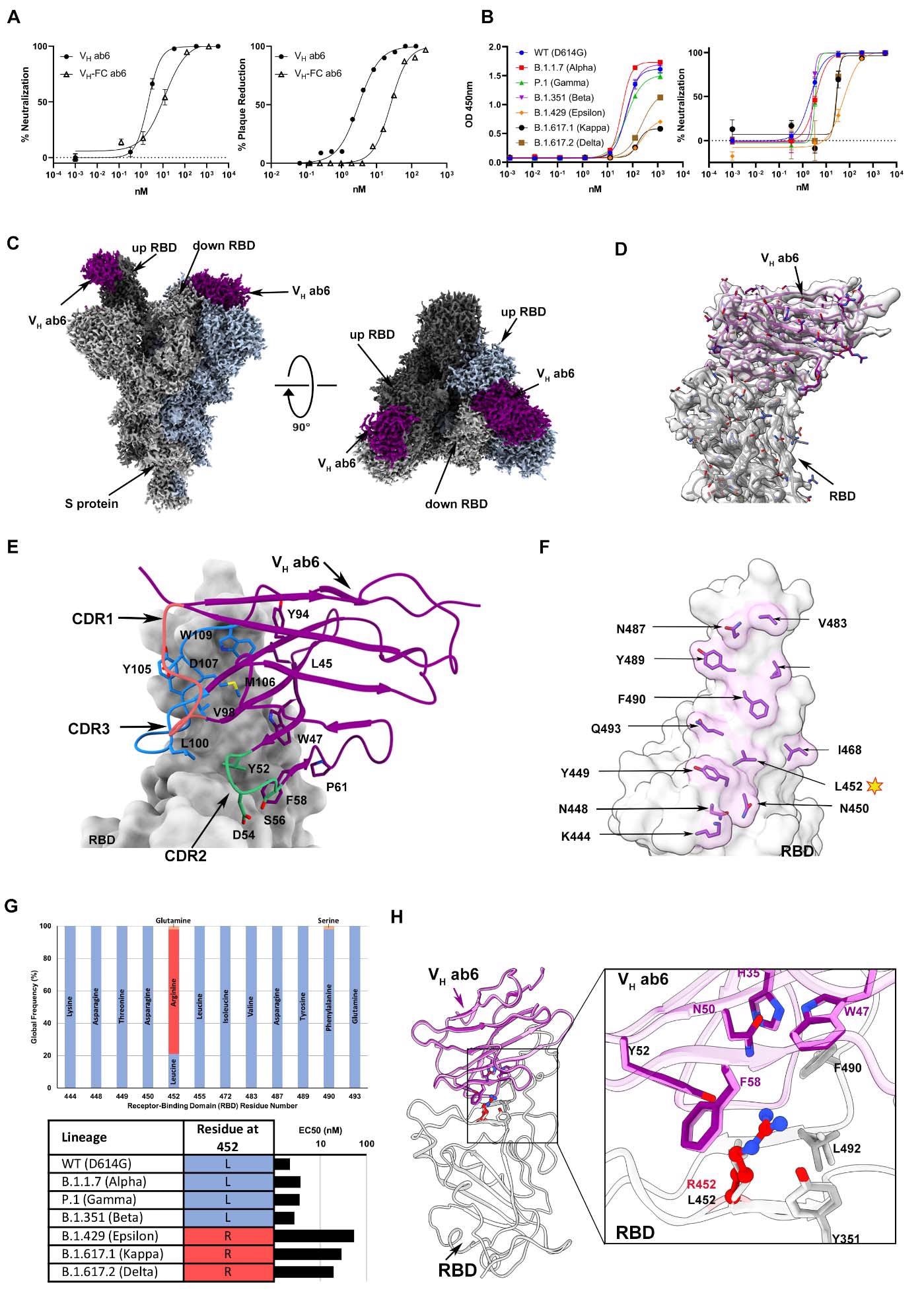 Ab6 broadly neutralizes SARS-CoV-2 variants via a largely conserved molecular epitope. (A) Pseudovirus neutralization (left) and authentic SARS-CoV-2 plaque reduction neutralization test - PRNT (right) by VH ab6 and VH-Fc ab6 against wild-type lineages. The pseudovirus neutralization (left) utilized the D614G wild-type, whereas the PRNT (right) utilized the USA-WA1/2020 isolate. (B) ELISA S protein binding (left) and pseudovirus neutralization (right) of SARS-CoV-2 variants by VH ab6. (C) 2.4Å global cryoEM density map of VH ab6 bound to wild-type S protein. Density corresponding to S protein protomers and ab6 are shown in grayscale and navy blue, respectively. (D) Focusrefined atomic model of ab6 bound to the wild-type (D614G) S protein RBD. (E) Analysis of ab6 contact zones. The RBD and ab6 are shown as a gray molecular surface and colorized cartoon representation, respectively. The ab6 scaffold is depicted in purple and the three complementarity determining regions (CDRs) of ab6 are coloured as follows: CDR1 - red; CDR2 - green; CDR3 - blue. (F) Footprint of ab6 mapped onto the wild-type S protein RBD. The sidechains of footprint residues are shown and colorized in purple. (G) (Top) Global frequency of residue identity within the ab6 footprint in GISAID deposited sequences as of August 1 st , 2021. (Bottom) Residue identity at position 452 within SARS-CoV-2 variants and associated VH ab6 EC50 measured via pseudoviral neutralization assays depicted on a logarithmic scale. (H) Focused view superpositions of the cryoEM-derived atomic model of the Epsilon (B.1.429) and wild-type (D614G) S proteins bound to VH ab6. Epsilon and wildtype RBD’s are coloured light and dark grey respectively, while purple and pink models refer to ab6-WT and ab6-Epsilon, respectively. The R452 mutation is highlighted in red. Pseudoviral neutralization experiments and ELISAs were performed in triplicate, error bars denote the standard
Ab6 broadly neutralizes SARS-CoV-2 variants via a largely conserved molecular epitope. (A) Pseudovirus neutralization (left) and authentic SARS-CoV-2 plaque reduction neutralization test - PRNT (right) by VH ab6 and VH-Fc ab6 against wild-type lineages. The pseudovirus neutralization (left) utilized the D614G wild-type, whereas the PRNT (right) utilized the USA-WA1/2020 isolate. (B) ELISA S protein binding (left) and pseudovirus neutralization (right) of SARS-CoV-2 variants by VH ab6. (C) 2.4Å global cryoEM density map of VH ab6 bound to wild-type S protein. Density corresponding to S protein protomers and ab6 are shown in grayscale and navy blue, respectively. (D) Focusrefined atomic model of ab6 bound to the wild-type (D614G) S protein RBD. (E) Analysis of ab6 contact zones. The RBD and ab6 are shown as a gray molecular surface and colorized cartoon representation, respectively. The ab6 scaffold is depicted in purple and the three complementarity determining regions (CDRs) of ab6 are coloured as follows: CDR1 - red; CDR2 - green; CDR3 - blue. (F) Footprint of ab6 mapped onto the wild-type S protein RBD. The sidechains of footprint residues are shown and colorized in purple. (G) (Top) Global frequency of residue identity within the ab6 footprint in GISAID deposited sequences as of August 1 st , 2021. (Bottom) Residue identity at position 452 within SARS-CoV-2 variants and associated VH ab6 EC50 measured via pseudoviral neutralization assays depicted on a logarithmic scale. (H) Focused view superpositions of the cryoEM-derived atomic model of the Epsilon (B.1.429) and wild-type (D614G) S proteins bound to VH ab6. Epsilon and wildtype RBD’s are coloured light and dark grey respectively, while purple and pink models refer to ab6-WT and ab6-Epsilon, respectively. The R452 mutation is highlighted in red. Pseudoviral neutralization experiments and ELISAs were performed in triplicate, error bars denote the standard
Spike Variants Escape RBD/NTD-Targeting Antibodies
The researchers used antibodies that target the RBD, one from each class of antibody, plus a highly neutralizing antibody S2M11, that binds two adjacent RBDs. Two anti-NTD antibodies, 4-8 and 4A8 were also used to examine NTD mutational impacts for each spike variant.
When the degree of antibody binding was measured and compared quantitatively with the degree of neutralization, the results showed that the cross-reactive SARS-CoV antibodies S309 and CR3022 continued to show intact binding across all spike variants. This was because their epitopes lie outside the RBD region affected by VOC mutations.
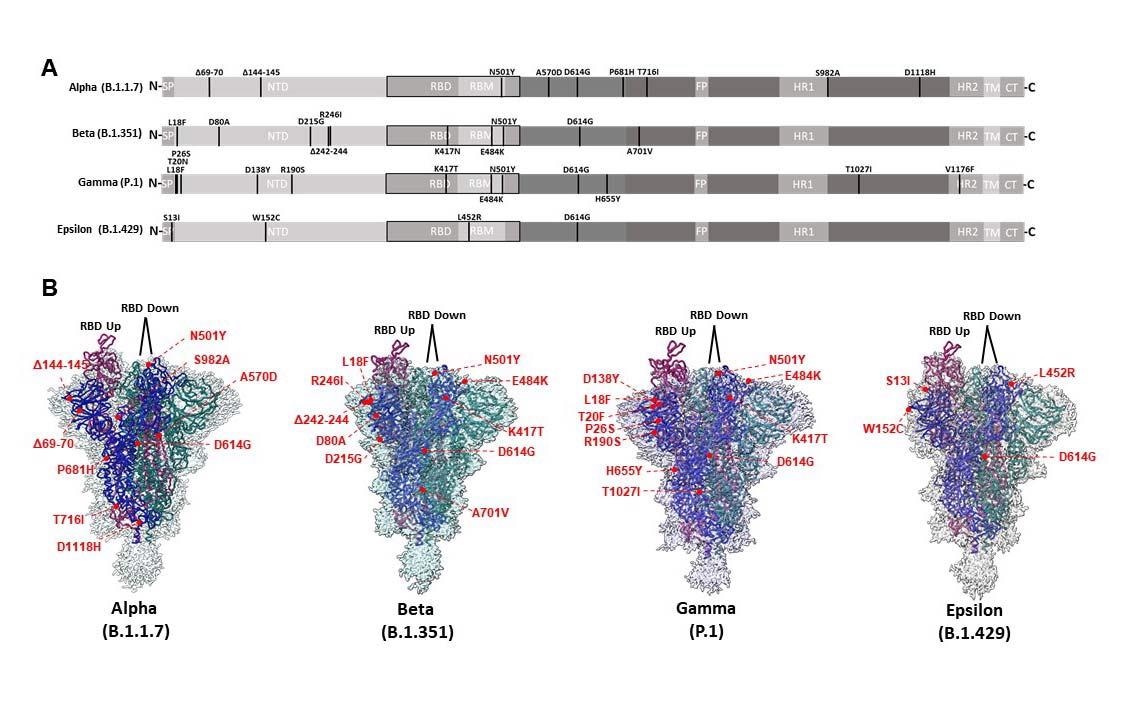
CryoEM structures of Alpha, Beta, Gamma, and Epsilon spike glycoproteins. (A) Linear schematic depicting mutations within variant S proteins (SP: Signal peptide, NTD: N-terminal domain, RBD: Receptor binding domain, RBM: Receptor binding motif, FP: Fusion peptide, HR1: Heptad repeat 1, HR2: Heptad repeat 2, TM: Transmembrane, CT: Cytoplasmic tail). (B) Global cryoEM maps and models for the Alpha (2.56 Å), Beta (2.56 Å), Gamma (2.25 Å), and Epsilon (2.4 Å) variant S proteins. Mutational positions are indicated and labeled in red.
Beta and Gamma variants showed drastic resistance to ab1 neutralization since both possess K417/T mutations. Both also resisted ab8 activity due to the presence of E484K mutations. The Epsilon spike variant showed lower but still present neutralization with S2M11, like ab6.
Further analysis showed that with Epsilon-ab6, R452 projected into the antibody-RBD interface, but with S2M11, it pointed away from the binding interface. If it is remembered that the mutations at this position, L452R, occur within a sidechain that fits within the epitope of S2M11, causing steric hindrance and repulsive charge effects, the likely increase in interaction energy may explain the weakening of neutralization potency.
Antibody binding to the RBD, with 4A8 and 4-8 antibodies, was also observed to escape with variants containing mutations near or in the antibody epitopes, supporting earlier reports that antigenic loops in the Alpha, Beta, and Epsilon NTDs were rearranged. This remodeling occurred as a direct or allosteric result of the NTD mutations. Examples include W152C in the Epsilon NTD, and the Beta Δ242-245 and R261I, which fall within the epitopes of one or both of these antibodies.
With polyclonal serum samples, binding antibodies to the wild-type RBD and NTD correlated with levels of neutralizing antibodies to these regions, showing their importance in viral neutralization by antibodies. This was impaired for all variants of the spike.
"These results highlight the role of mutations within the NTD and RBD of variant spikes in driving evasion of SARS-CoV-2 directed monoclonal and polyclonal antibodies, providing a basis to evaluate the broad mutational tolerance exhibited by S309 and ab6."
The mutational clusters within the spike variants also affect ACE2 binding by the RBD, albeit slightly, which may also contribute to the increased infectivity. When the structural impact of the mutations was examined for each spike, they found varying effects.
For instance, A570D and S982A mutations in the Alpha VOC affected RBD positioning, with different positions and interactions depending on the "up" or "down" conformation of the adjacent RBD protomer. This ranges from hydrogen bonding to salt bridge formation. The latter, between K852-D570, stabilizes the RBD "up" protomer, while S982 results in destabilizing the "RBD down" protomers.
The Alpha D1118H mutation increases the number of contacts between adjacent RBD protomers, further stabilizing the spike in trimeric form.
Similar interactions in Beta and Gamma variants show tight packing effects or more robust salt bridge networks, stabilizing these trimers. However, mutations outside the RBD failed to affect the position of residues in contact with the ACE2 receptor interface allosterically.
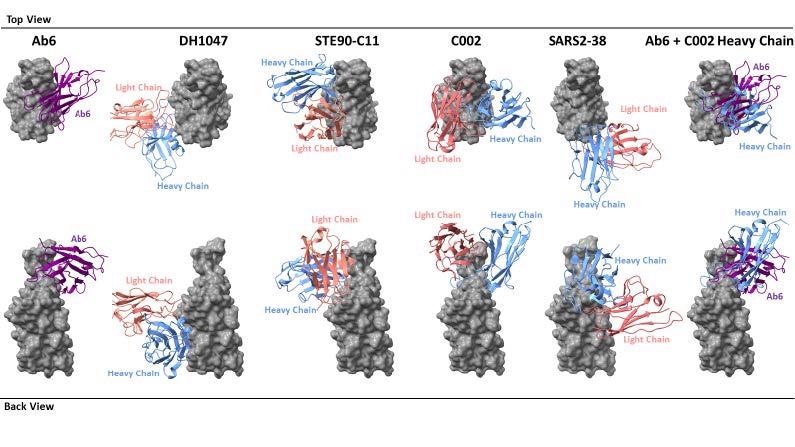
Footprint comparison between ab6 and selected RBD-directed antibodies. The RBD is depicted as a grey molecular surface and antibodies are depicted as colorized cartoon models. The following PDB files were utilized: 7LD1 (DH1047), 7B3O (STE90-C11), 7K8T (C002), 7MKM (SARS2-38). The RBD model from the ab6-RBD complex is shown for all antibody complexes for ease of visualization. For superpositions, structures were aligned using the RBD.
What Are the Implications?
The antibody epitope described here is at a distance from most mutations in the VOCs, and this antibody can thus neutralize most VOCs. This could allow the rational therapeutic design to target viral proteins found in different variants.
The analysis shows that most of the spike variants show little structural changes in the RBD, indicating the essentiality of the latter for ACE2 binding. Conversely, the mutations found in the variant NTDs resulted in the rearrangement of the NTD antigenic loops, enabling immune evasion. "These contrasting structural effects between variant NTD and RBD mutations likely arise due to different functional requirements and selective pressures between these domains."
The NTD effects show that antibody escape drives most NTD mutations, including A570D and S982A mutations in the RBD of the Alpha variant. Conversely, the D1118H and T1027I mutations within the S2 subunit of the Alpha and Gamma spikes, respectively, increase the strength of the salt bridges between adjacent protomers, stabilizing the spike variants. The mutations in different regions thus produce different effects on spike function and mirror the varying drivers of mutations in each region.
However, neutralizing epitopes are primarily located within the RBD and NTD, as shown by the strong correlation between RBD and NTD binding and neutralizing epitopes in the wild-type spike. Lower degrees of correlation are observable with variant spikes, but neutralization is still preserved. This indicates that neutralizing epitopes are preserved in the RBD, as shown by the intact potency of S309, an antibody elicited by SARS-CoV, against all variant spikes.
The study also demonstrates the presence of a novel RBD epitope that binds to a broadly neutralizing antibody against all VOC spikes, including the Delta variant that is currently dominant in most of the world. This epitope has been conserved across all variants, with only the L452R mutation having been detected so far. The ab6 antibody has been found to bind to this epitope even with this mutation, though the latter causes a decreased neutralizing potency.
The detailed structure of the spike as described here could also help develop better therapeutics and explain why these variants are fitter than the wild-type, accounting for their dominant place, in terms of better ACE2 affinity and antibody escape.
Recently, it was shown that there are seven types of RBD binding antibodies and that broadly neutralizing antibodies bind to cryptic epitopes inside the RBD, while antibodies that do not compete with the ACE2 receptor bind to the outside of the RBD. The antibody in this study, ab6, is of the first class, binding to the inner RBD face, making contact with the receptor-binding motif and thus competing with ACE2 for binding.
Structurally, it overlaps with a representative of the RBD-4 class of antibody, C002, though with different binding modes. The latter was isolated from a convalescent COVID-19 patient, which indicates that the binding epitope for ab6 is recognized by natural antibodies also. "This evidence further supports the potential value of focus on the ab6 binding epitope for future therapeutic design."

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources