Interesting research decrypts a recurring sequence in antibodies against severe acute respiratory syndrome 2 (SARS-CoV-2) and identifies it to be a board neutralizing antibody that targets a conserved epitope on the SARS-CoV-2 receptor-binding domain (RBD) of the spike protein.
The identified recurrent sequence, the YYDRxG motif, is identified on many antibodies and broadly neutralizes SARS-CoV-2 variants and SARS-CoV.
This study, recently published on the preprint server bioRxiv*, suggests that the identified motif represents a common convergent solution for the immune system to counteract sarbecoviruses. Significantly, it is also useful as an epitope targeting strategy for pan-sarbecovirus vaccines and antibody therapeutics.
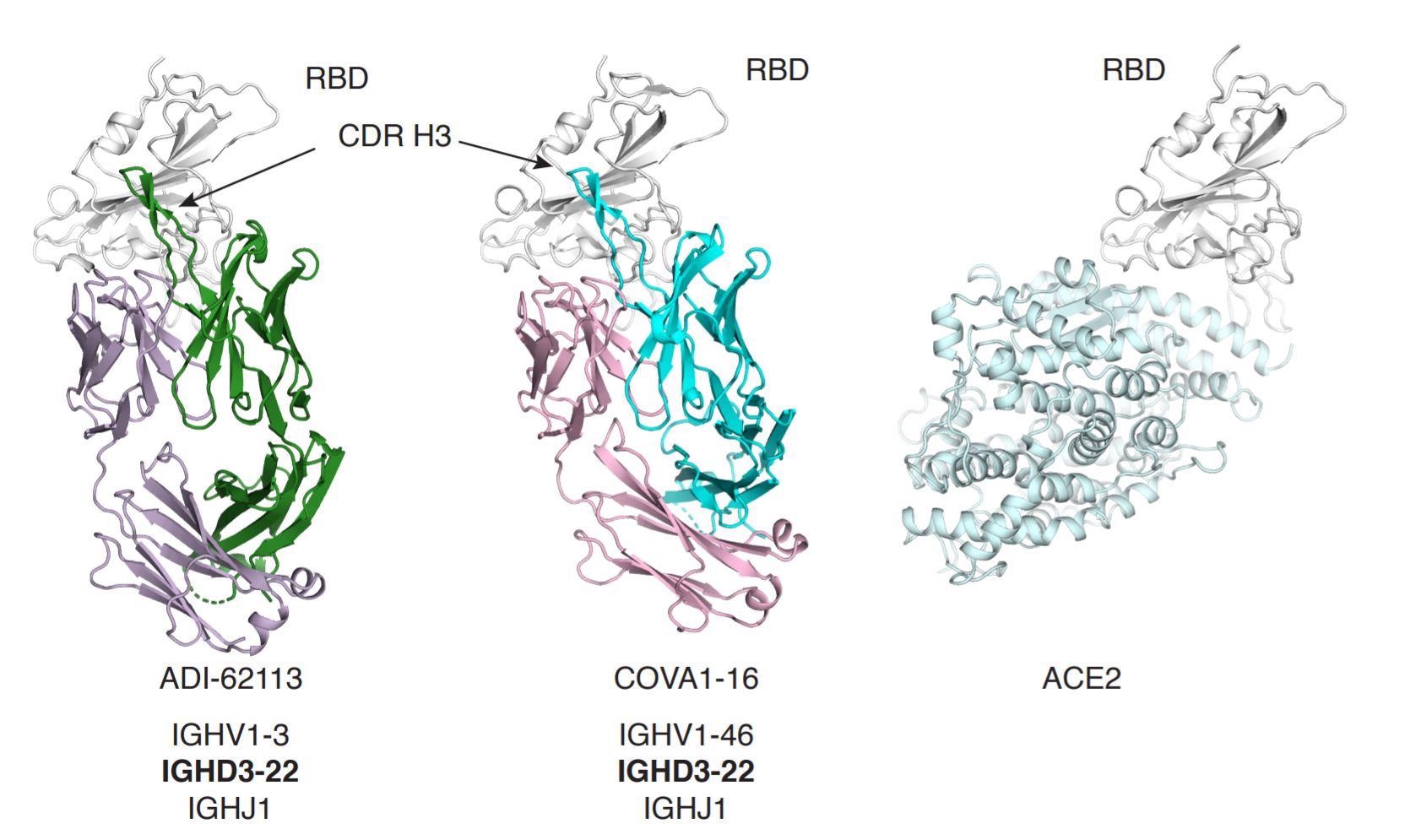
Structural comparison of ADI-62113 and COVA1-16

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Introduction
Zoonotic spillovers have affected human lives and farm animals. The ongoing coronavirus disease 2019 (COVID-19) pandemic, caused by SARS-CoV-2, has so far affected over 273.9 million individuals and caused over 5.4 million deaths across the globe. Now the virus has been rapidly mutating, giving rise to emerging variants - prolonging the fight to control the COVID-19 pandemic.
The viral antigenic drift causing enhanced infectivity and immunity escape is a major challenge in overcoming the current pandemic. While a majority of the potent antibodies target the RBD of the spike protein of SARS-CoV-2, mutations in these regions may render these antibodies ineffective.
However, a few broadly neutralizing antibodies (bnAbs) can target more conserved surfaces on the viral spike protein to neutralize circulating variants of concern (VOC), variants of interest (VOI), and other SARS-related viruses in the sarbecovirus family.
Therefore, to fight the current COVID-19 pandemic, we urgently need to identify and characterize such cross-neutralizing antibodies, with a common motif that can be used to develop an epitope targeting strategy.
Study findings
The antibody, ADI-62113, is a potent cross-neutralizing antibody which was isolated from a COVID-19 patient. However, its genes, the immunoglobulin heavy variable gene IGHV1-3 (heavy chain) and kappa variable gene IGKV1-33 (light chain) have not been reported in other SARS-CoV-2 cross-neutralizing antibodies.
Expressing various sarbecovirus RBDs on the surface of yeast, the researchers characterized the binding kinetics with the antibody ADI-62113 and found that it has a high affinity for a broad spectrum of sarbecoviruses. The antibody also binds to a highly conserved site (CR3022) on the SARS-CoV-2 RBD.
Notably, the researchers found that the YYDRxG motif is shared between antibodies ADI-62113 and COVA1-16, revealing a highly similar binding mode to the SARS-CoV-2 RBD as first observed in COVA1-16.
Furthermore, the researchers reported that the YYDRxG motif is located at the tip of CDR H3 and precedes a G1 β-bulge in the descending strand of the hairpin structure. They discussed the configuration, structural stability, and specificity of CDR H3 in its interaction with the RBD.
As the conserved CR3022 site extensively interacts with the motif on the RBD, and the CDR H3 local structure is stabilized by a β-bulge, the YYDRxG pattern is a desirable feature for specific RBD recognition in strategizing against the virus. The researchers noted that this pattern is more extensive than for the recurrent YYD sequences in HIV V2-apex targeting bnAbs.
Searching over 205,000 antibody sequences, the researchers found that 153 antibodies had a YYDRxG pattern in their CDR H3 with most of these YYDRxG motifs being found to be encoded by the IGHD3-22 gene. The researchers presented the Reading frames (RFs) and sequence alignment of IGHD3-22; the data indicates sites where somatic hypermutations are often found in YYDRxG neutralizing antibodies against SARS-CoV-2.
The researchers found that the YYDRxG motif is associated with potent and broad anti-SARS-CoV-2 antibodies. When computationally searching for this YYDRxG pattern, the researchers identified and verified many such antibodies that are potent neutralizers of SARS-CoV-2, with a wide breadth of activity against the emerging and circulating viruses (VOCs, and SARS-CoV). They specifically identified 100 antibodies with the conserved YYDRxG motif exclusively encoded by the IGHD3-22 gene.
This is a time- and effort-saving attempt in finding a potent and broadly neutralizing antibody as opposed to the conventional, resource-limiting experimental and structural characterization for antibodies.
Significantly, the researchers suggested that because these antibodies containing a YYDRxG feature are elicited by both natural infection and vaccination, looking for these signature sequences in the serum may serve as biomarkers to evaluate the vaccine breadth and guide the rational design of next-generation vaccines.
The YYDRxG pattern search identifies broad and potent antibodies to sarbecoviruses
Conclusion
Researchers structurally characterized a cross-neutralizing antibody, ADI-62113, and identified a recurrent motif, YYDRxG, a hexapeptide encoded by IGHD3-22 in heavy-chain complementarity-determining region 3 (CDR H3). Then, they conducted a computational search for the YYDRxG sequence pattern in publicly available database sequences of antibodies and identified 100 antibodies with broad neutralization activity against SARS-CoV-2 variants and SARS-CoV.
This study identified a potent recurrent motif that can be used in broadly neutralizing antibodies, as ‘a common convergent solution for the human humoral immune system to counteract sarbecoviruses.’
Importantly, this study shows that cross-neutralizing, bnAbs, can be rapidly identified from their sequence alone.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
A recurring YYDRxG pattern in broadly neutralizing antibodies to a conserved site on SARS-CoV-2, variants of concern, and related viruses. Hejun Liu, Chengzi I. Kaku, Ge Song, Meng Yuan, Raiees Andrabi, Dennis R. Burton, Laura M. Walker, Ian A. Wilson. bioRxiv 2021.12.15.472864; DOI: https://doi.org/10.1101/2021.12.15.472864
- Peer reviewed and published scientific report.
Liu, Hejun, Chengzi I. Kaku, Ge Song, Meng Yuan, Raiees Andrabi, Dennis R. Burton, Laura M. Walker, and Ian A. Wilson. 2022. “Human Antibodies to SARS-CoV-2 with a Recurring YYDRxG Motif Retain Binding and Neutralization to Variants of Concern Including Omicron.” Communications Biology 5 (1): 1–13. https://doi.org/10.1038/s42003-022-03700-6. https://www.nature.com/articles/s42003-022-03700-6.