A team of scientists from Canada have recently characterized a panel of nanobodies against the spike protein of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The nanobodies have shown high epitope binding affinity and neutralizing efficacy against a wide variety of SARS-CoV-2 variants of concern (VOCs). The study is currently available on the bioRxiv* preprint server.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
The coronavirus disease 2019 (COVID-19) pandemic caused by SARS-CoV-2 has put a tremendous burden on the global healthcare system, with more than 275 million infections and 5.5 million deaths worldwide. Despite the development of potential vaccines and therapeutic monoclonal antibodies against COVID-19, a continuous uprise in pandemic trajectory has been observed globally particularly because of the emergence of mutated viral variants with improved immune fitness.
Besides monoclonal antibodies, single-domain nanobodies have shown considerable potency as a therapeutic intervention against COVID-19. Nanobodies are recombinant variable domains of heavy-chain-only antibodies found in camelids. The high aerosol-ability of nanobodies makes them suitable candidates for inhalation-based delivery. Moreover, nanobodies can be assembled to form multimeric constructs that exhibit a high cross-reactivity and neutralizing ability against a variety of VOCs.
In the current study, the scientists have characterized a panel of 37 anti-SARS-CoV-2 spike nanobodies as both monovalent and IgG Fc-fused bivalent modalities.
Epitope-binding abilities of nanobodies
The analysis of binding characteristics showed that both monovalent and bivalent nanobodies have high affinity against the spike protein, S1 and S2 domains, receptor-binding domain (RBD), and N-terminal domain (NTD) of the original Wuhan strain of SARS-CoV-2. In addition, the majority of bivalent nanobodies showed high cross-reactivity against the spike protein of alpha, beta, gamma, delta, and kappa variants of SARS-CoV-2. Similarly, most of the monovalent nanobodies showed high cross-reactivity against pangolin coronavirus, with some showing cross-reactivity against SARS-CoV. Monovalent nanobodies targeting the S2, RBD, and NTD domains of SARS-CoV-2 showed a broad range of cross-reactivity.
Importantly, most of the monovalent nanobodies showed considerable stability against high temperature and aerosolization. Similarly, bivalent nanobodies showed high serum stability and persistence suitable for conducting animal experiments.
Neutralizing efficacy of nanobodies
The virus neutralization assay findings revealed that a large number of monovalent nanobodies, especially anti-RBD nanobodies, are able to neutralize SARS-CoV-2. However, the proportion of bivalent nanobodies with neutralizing ability was higher than monovalent nanobodies.
The neutralization efficacy of all anti-RBD monovalent and bivalent nanobodies and some anti-NTD bivalent nanobodies against tested VOCs were determined in the study. The findings revealed that the nanobodies capable of effectively neutralizing the wild-type virus and the alpha variant exhibit lower neutralizing efficacy against the tested variants.
A total of 12 out of 20 tested bivalent nanobodies exhibited high efficacy in neutralizing the delta variant. Of these nanobodies, 9 were capable of neutralizing all tested variants as well as SARS-CoV.
Reformatting of monovalent nanobodies to bivalent nanobodies caused a significant improvement in neutralizing efficacy. The improvement was highest for anti-RBD nanobodies (2 – 100-fold). Reformatting even transformed some non-neutralizing monovalent nanobodies into potent neutralizers.
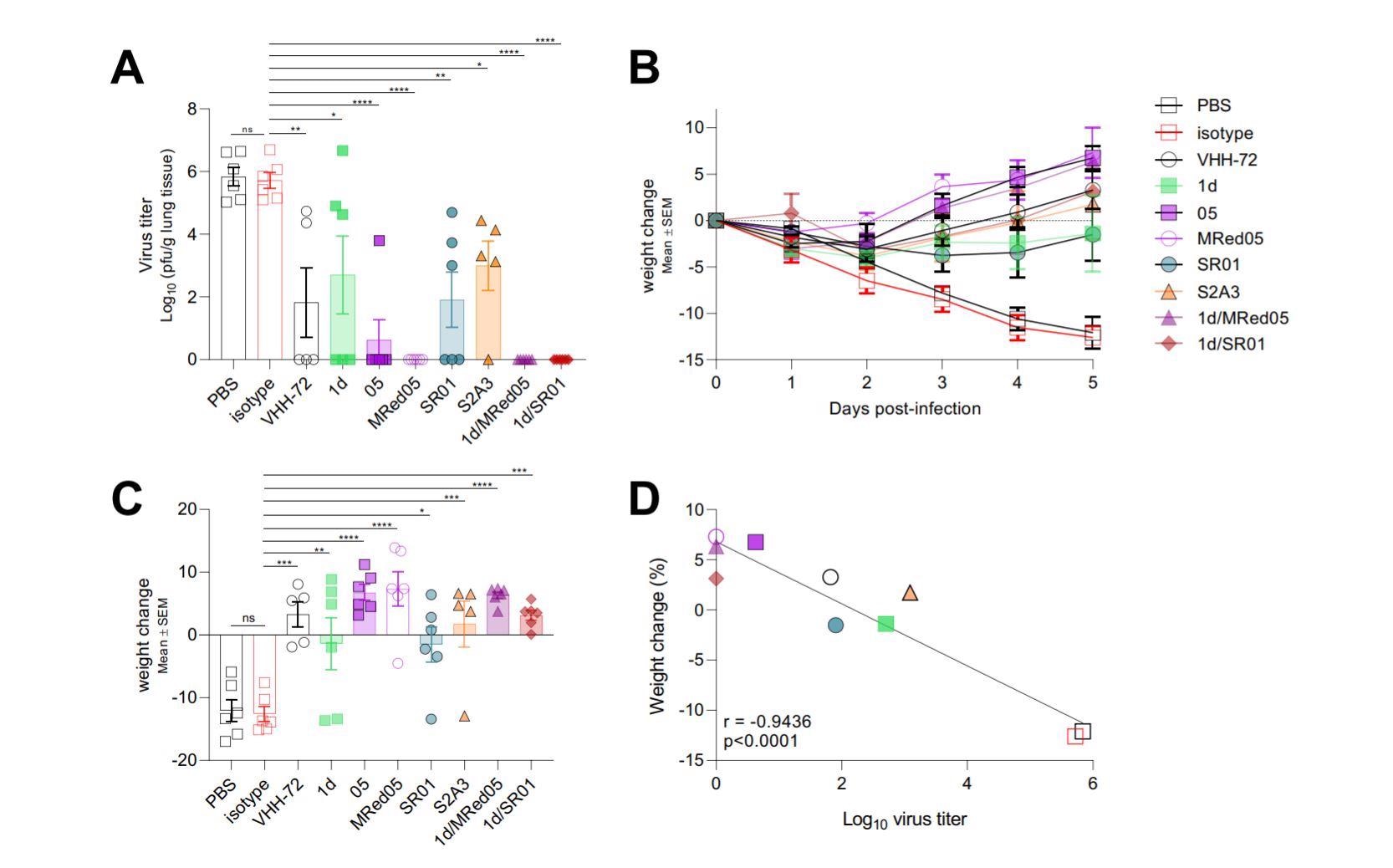
VHH-Fcs showed strong protective efficacy in hamsters challenged with SARS-CoV-2.
Epitope characterization
A total of 14 unique epitope bins were identified for 33 tested monovalent nanobodies. Of these epitope bins, 6 were for anti-RBD nanobodies, 3 for anti-NTD nanobodies, and 5 for anti-S2 nanobodies.
Regarding conformational arrangements of epitope bins, a range of binding patterns were identified. Moreover, a correlation between stabilizations spanning mutations in VOCs and the loss of neutralization was observed in the study.
Therapeutic efficacy of nanobodies
The therapeutic efficacy of five bivalent nanobodies (3 anti-RBD, 1 anti-NTD, and 1 anti-S2 nanobodies) was tested in hamsters infected with SARS-CoV-2.
Compared to untreated control animals, nanobody-treated animals showed significantly lower lung viral titers and improved body weight loss after 5 days of infection. The treatment with a combination of two nanobodies caused the highest reduction in viral titers.
The immunohistochemistry study findings revealed that the nanobodies effectively reduce SARS-CoV-2-induced inflammatory responses, lung apoptosis, and infiltration of inflammatory immune cells in the lungs.
Study significance
The study characterizes a panel of nanobodies with good neutralizing efficacy against SARS-CoV-2 VOCs. Given the therapeutic efficacy in animals, the scientists suggest that these nanobodies can be used as potential therapeutics against COVID-19.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources