
 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
In November 2021, the fifth SARS-CoV-2 VOC known as the Omicron variant was identified and found to have 26 to 32 spike protein mutations. The surveillance of new SARS-CoV-2 variants has been achieved through the sequencing of clinical samples; however, the techniques used for these purposes are often associated with high operational costs and specialized infrastructure requirements.
In light of these challenges, an alternative method of wastewater-based surveillance (WBS) has garnered attention due to its low cost and real-time unbiased insights into virus transmissibility for the entire population at the community level. In fact, WBS has already proven effective in assessing SARS-CoV-2 circulation in several different countries, as well as for detecting the spread of new SARS-CoV-2 strains.
Despite the utility of WBS, this surveillance approach is associated with poor sensitivity in identifying low-frequency variants. Furthermore, there remains a lack of quantitative modeling for WBS that would be required for its widespread application.
About the study
For the clinical diagnosis of the coronavirus disease 2019 (COVID-19), RT-qPCR is considered the gold standard for detecting the presence of SARS-CoV-2.
In an effort to expand the application of this analytical technique, the researchers of the current study developed a new set of AS RT-qPCR primers to detect and quantify the Omicron variant from wastewater using two approaches. The first approach is a new assay targeting the Q493R, G496S, and Q498R loci found in the majority of Omicron mutations,, whereas the second approach involves targeting the HV69-70del mutation found in both the Omicron and Alpha variants.
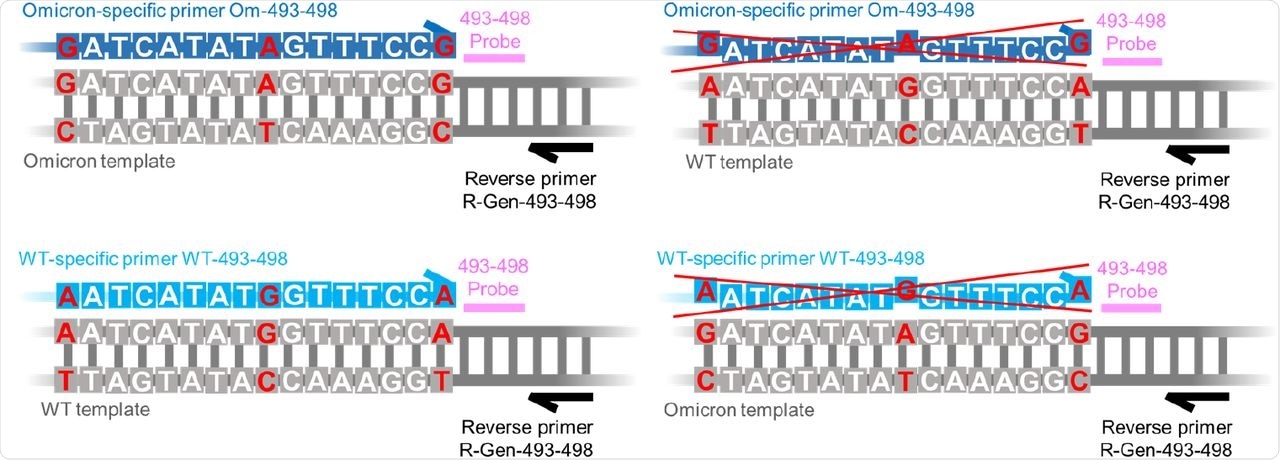
Schematic of our panel of AS RT-qPCR assays.
Study findings
The current study assessed the specificity for wild-type (WT) and mutant targets at locus 493-498 by screening the synthetic DNA containing WT and Omicron sequences at these targeted loci by qPCR reactions.
The assay demonstrated high specificity in identifying the WT and Omicron variants in wastewater samples. These properties were demonstrated by the lack of cross-reactivity observed in the WT DNA and Omicron variant genotypes at up to 103 and 104 copies of DNA of the opposite genotype, respectively.
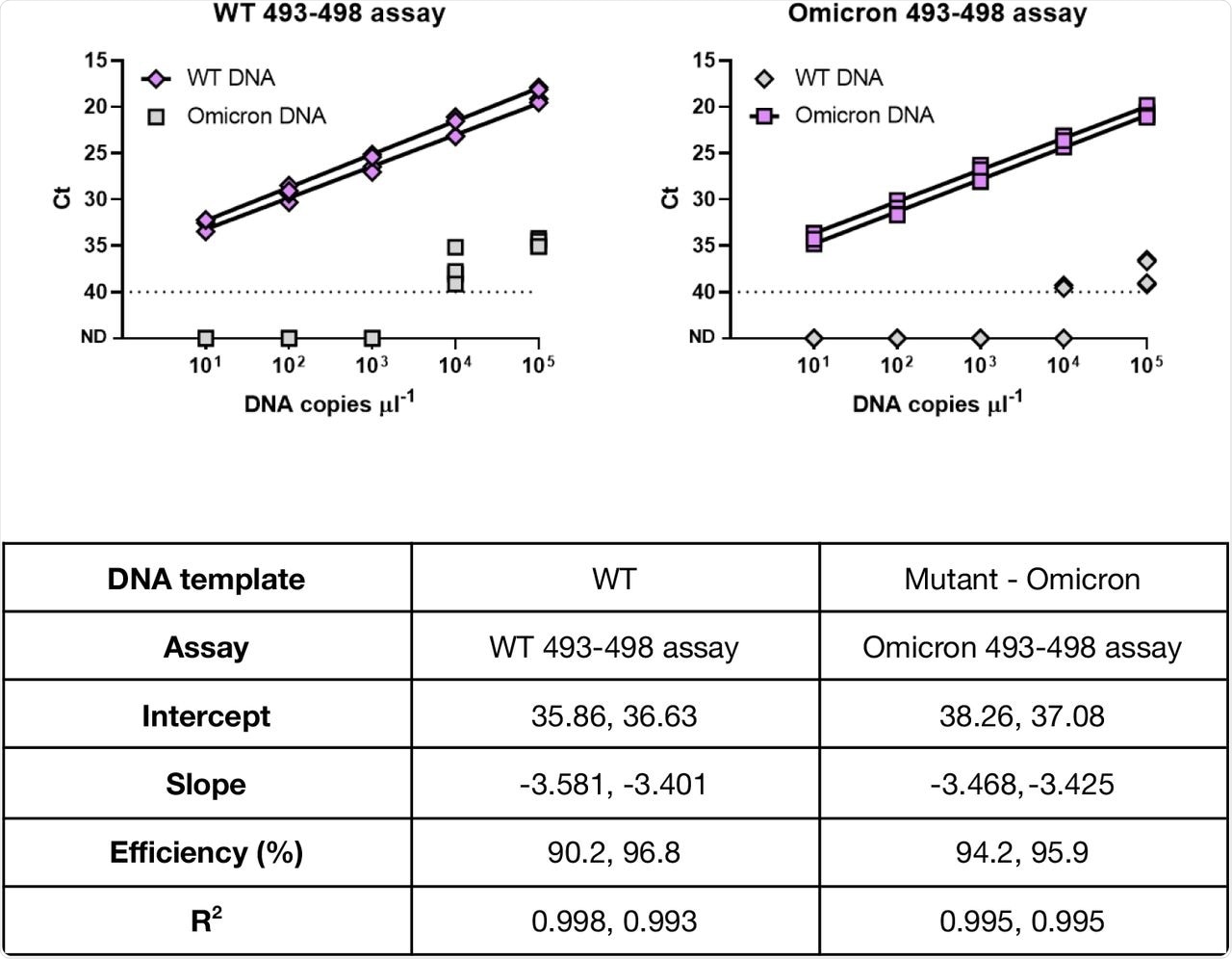
Specificity and cross-reactivity of AS RT-qPCR primers against the WT and the Omicron DNA sequences.
The researchers further evaluated the twist synthetic SARS-CoV-2 ribonucleic acid (RNA) control and found the amplification efficiency from 90% to 94% for DNA sequences representative of Omicron and WT assay for WT sequences. The sensitivity of this assay was equivalent to the widely used United States Centers for Disease Control and Prevention (CDC) assay.
The second approach assessed in this study was previously to detect the Alpha variant and was used in the present study for detecting the Omicron variant. Herein, the 69-70 deletion targeting assay reported high specificity and no cross-reaction up to 105 copies of RNA of the opposite WT genotype. The sensitivity of this 69-70 del targeting assay was comparable to the widely accepted CDC N1/N2 assays.
Limitations and conclusion
The present assay was validated using the synthetic DNA sequences of WT and Omicron variant and against WT RNA; however, the RNA for the Omicron variant was not available for this study. Another limitation was that the authors did not validate the Omicron variant sequences in real wastewater samples.
The researchers shared this assay and its initial validation findings to demonstrate the benefits of RT-qPCR for WBS purposes. According to the authors, this novel assay would serve as an important tool for tracking the transmissibility of the Omicron variant at the community level at a low cost.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Lee, W. L., Gu, X., Armas, F., et al. (2021). Quantitative detection of SARS-CoV-2 Omicron variant in wastewater through allele-specific RT-qPCR. medRxiv. doi:10.1101/2021.12.21.21268077. https://www.medrxiv.org/content/10.1101/2021.12.21.21268077v1.full-text.
- Peer reviewed and published scientific report.
Lee, Wei Lin, Maxim Imakaev, Federica Armas, Kyle A. McElroy, Xiaoqiong Gu, Claire Duvallet, Franciscus Chandra, et al. 2021. “Quantitative SARS-CoV-2 Alpha Variant B.1.1.7 Tracking in Wastewater by Allele-Specific RT-QPCR.” Environmental Science & Technology Letters 8 (8): 675–82. https://doi.org/10.1021/acs.estlett.1c00375. https://pubs.acs.org/doi/10.1021/acs.estlett.1c00375.