The Omicron variant of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus contains extensive sequence changes relative to earlier SARS-CoV-2 variants that, to date, have unknown effects on its infectivity and vaccine-induced immunity. As of December 23, 2021, SARS-CoV-2, the causal agent of the coronavirus disease 2019 (COVID-19), has infected over 278 million worldwide claimed almost 5.4 million lives.
Study: Omicron mutations enhance infectivity and reduce antibody neutralization of SARS-CoV-2 virus-like particles. Image Credit: Fit Ztudio / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
Gaining a deeper understanding of the molecular determinants of SARS-CoV-2 viral fitness is essential for effective vaccine and therapeutic development. This is even more important in the current situation, owing to the emergence of new variants like the Delta and Omicron strains.
Prior research has shown that vesicular stomatitis virus (VSV) and lentiviruses pseudotyped with the SARS-CoV-2 S protein could aid in the evaluation of cell binding and entry into host cells through the angiotensin-converting enzyme 2 (ACE2) and TMPRSS2 receptors. Despite their utility, these pseudoviruses are unable to determine the effects of mutations outside the S gene.
About the study
In the current study, researchers developed SC2-VLPs consisting of the S, N, membrane (M), and envelope (E) structural proteins. The particles also consist of signal-containing messenger ribonucleic acid (mRNA). These signals form the RNA-loaded capsids that are responsible for S protein-dependent cell transduction.
This system appears to be representative of the impact of mutations in structural proteins that are observed in live-virus infections. Further, the procedure used in the current study enables the quick testing of SARS-CoV-2 structural gene variants for their impact on antibody or serum neutralization, as well as infection efficiency.
Study findings
To compare the effects of different structural gene variants on infectivity, the researchers used the structural genes from the SARS-CoV-2 B.1 strain as a point of reference. They observed that relative to S-B.1, the Delta variant produced VLPs that were less than half as infectious, while the Omicron variant generated VLPs that were more than twice as infectious as the original virus. This result was not in line with previous observations of pseudotyped lentiviral particles, thus showing the increased entry for S-Delta and reduced entry for S-Omicron.
The scientists then compared the effects of N, M, or E viral variants on the infectivity of VLPs generated using B.1 genes. These studies were conducted in response to previous research demonstrating that the N gene had a pronounced influence on infectivity and RNA packaging efficiency.
Consistent with previous research, the current study also found that the N-Delta and N-Omicron variants generated VLPs with robust infectivity that was higher as compared to both the B.1 and B.1.1 variants.
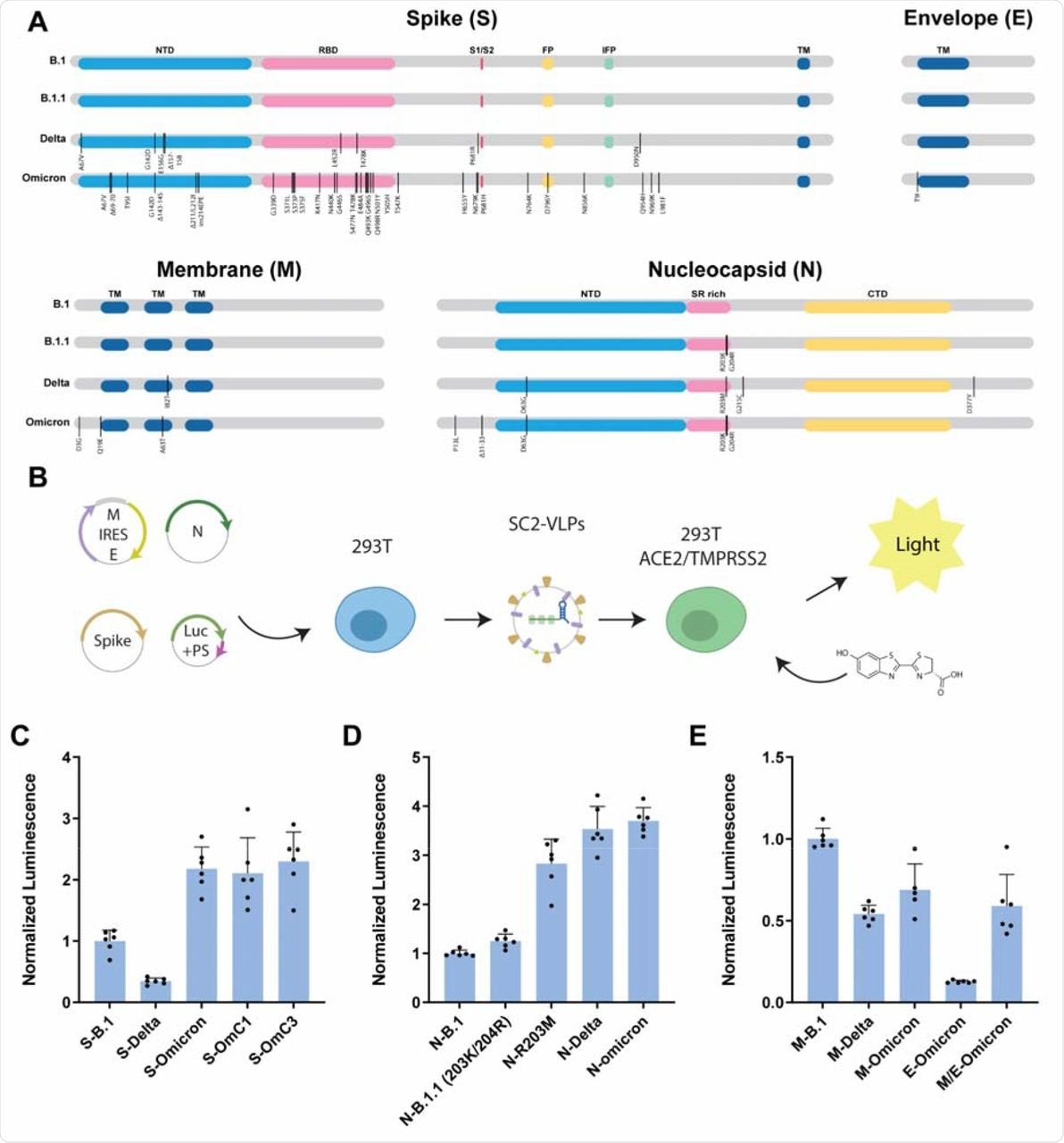
Omicron structural gene variants alter infectivity of SC2-VLPs.
The researchers then evaluated the VLP neutralization capability of antisera collected from 38 individuals four to six weeks post-vaccination or convalescent sera from unvaccinated COVID-19 survivors. The vaccines considered were Pfizer/BioNTech, Moderna, or Johnson & Johnson.
Preliminary investigation showed that sera from both Pfizer/BioNTech and Moderna vaccinated individuals yielded high neutralization titers as compared to sera obtained from Johnson & Johnson vaccinated individuals and convalescent patients. Testing the neutralization capacity of each patient's sera against VLPs from B.1, Delta, or Omicron viral variants revealed a pronounced decrease between 15-18-fold in potency against the Omicron variant, while the effect was intermediate for the other variants.
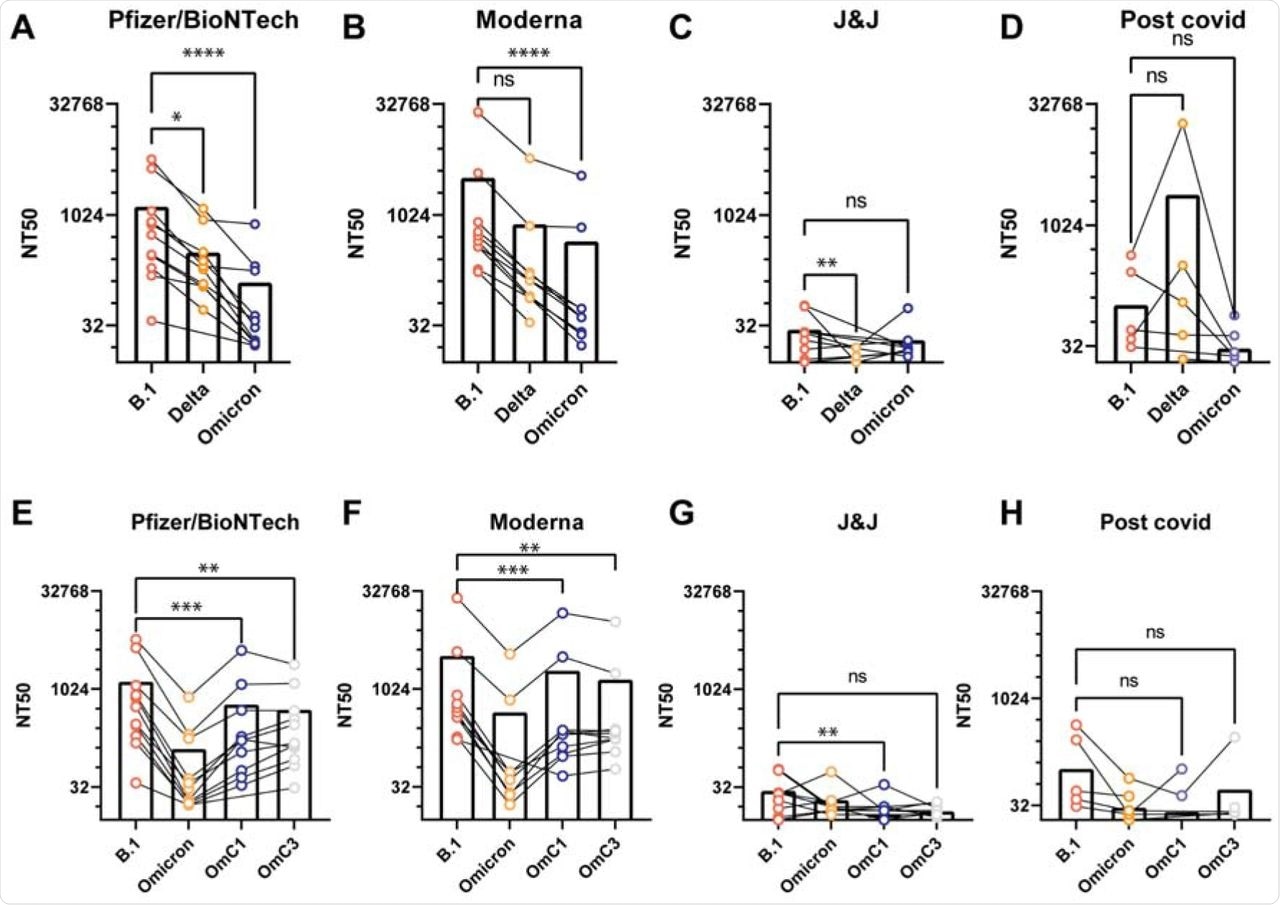
Antiserum neutralization of VLPs generated with different S genes.
The researchers were also interested in determining the effect of the monoclonal antibodies of Casirivimab and Imdevimab against the Omicron variant. Both antibodies showed robust neutralization activity against B.1.1 and Delta VLPs; however, neither showed any activity against Omicron VLPs.
The failure of these monoclonal antibodies to neutralize Omicron S could be due to six mutations within the Omicron receptor-binding domain (RBD) including K417N, N440K, G446S, G496S, Q498R, and N501Y.
Conclusion
The current study showed that Omicron SC-VLPs have enhanced infectivity and are only weakly neutralized by vaccination, prior infection, or antibody therapeutics. The results presented in this study suggest that the Omicron variant is at least as efficient at assembly and cell entry as Delta.
A concerning implication is that the antibody response triggered by vaccination or previous infection could be limited in its ability to neutralize the Omicron variant, at least prior to boosting. Additionally, it is expected that some of the currently available monoclonal antibodies will not be useful in treating Omicron-infected patients.
Although the data presented here did not account for T-cell-based immunity by vaccination or prior infection, existing evidence suggests that mRNA vaccine boosters could enhance the likelihood of protection against Omicron infection.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Syed, M. A., Ciling, A., Khalid, M. M., et al. (2021) Omicron mutations enhance infectivity and reduce antibody neutralization of SARS-CoV-2 virus-like particles. medRxiv. doi:10.1101/2021.12.20.21268048. https://www.medrxiv.org/content/10.1101/2021.12.20.21268048v1.
- Peer reviewed and published scientific report.
Syed, Abdullah M., Alison Ciling, Taha Y. Taha, Irene P. Chen, Mir M. Khalid, Bharath Sreekumar, Pei-Yi Chen, et al. 2022. “Omicron Mutations Enhance Infectivity and Reduce Antibody Neutralization of SARS-CoV-2 Virus-like Particles.” Proceedings of the National Academy of Sciences 119 (31). https://doi.org/10.1073/pnas.2200592119. https://www.pnas.org/doi/full/10.1073/pnas.2200592119.