Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infects cells by binding to angiotensin-converting enzyme 2 (ACE2) sites on epithelial cells. This is mediated by the spike protein, which consists of two subunits, S1 and S2, that must be cleaved by a host protein before becoming functional. S1 contains a receptor binding site (RBD) responsible for ACE2 binding and cell entry, while S2 mediates membrane fusion. Mutations in the spike protein, often in the RBD, are some of the most significant changes seen in new variants. Researchers from the University of Colorado Anschutz Medical Campus have been investigating the changes imparted on the Gamma variant by three RBD mutations.
A preprint of the groups' study can be found on the bioRxiv* server.
 Study: SARS-CoV-2 comes up with a perfect recipe for disaster: Each mutation plays a specific role in the evolution of variants of concern. Image Credit: NIAID
Study: SARS-CoV-2 comes up with a perfect recipe for disaster: Each mutation plays a specific role in the evolution of variants of concern. Image Credit: NIAID

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The Study
The researchers first examined the effect of the three RBD mutations found in the Gamma variant on expression, stability, ACE2 binding, and antibody escape. Single, double, and triple RBD mutants of these three mutations, K417T, E484K and N501Y, were created. These were expressed in Expi293 cells alongside wild-type RBD, allowing them to move through the endoplasmic reticulum where and misfolded proteins can be removed. The mutations significantly altered the amount of protein secreted by the cells. K417T was found to increase the final titer by 70% compared to wild-type, and E484K decreased it by 60%. N501Y reduced expression by 20%. When the double and triple mutants were investigated, these effects were found to be additive. Mutants that included K417T almost always showed higher expression, with K417T/N501Y increasing expression by 40%, K417T/E484K, and the triple mutant increased expression by 10%. When E484K and N501Y were together, expression was decreased by 80%, and the protein could not be purified.
The scientists used circular dichroism, a form of spectroscopy, to investigate the effects of these mutations on the secondary and tertiary structure of the RBD; however, they found that none of the mutants showed significant differences in structure.
Thermal and chemical denaturation melts were used to investigate the stability of wild-type compared to the mutants. More stable proteins can withstand more deleterious mutations and persist in the viral pool longer. The thermal melts showed the Tm of wild-type was 56.1 +/- 0.7C. N501Y slightly decreased this, but not significantly. K417T increased the thermal stability to 59.3+/-0.2C, showing similar values for the double and triple mutations. E484K decreased the thermal stability to 52.3+/-0.5C. Once again, the changes in stability were found to be additive, with double mutants showing values that were roughly the average of the two initial mutations. The chemical denaturation was performed using urea and tested using fluorescence spectroscopy. Free energy change on unfolding for wild-type was shown to be 8.1kcal/mol. The same pattern was seen for thermal stability, with K417T increasing stability, while the other two mutations reduced it. However, unlike thermal stability, this effect does not seem to be additive, with the triple mutant showing the least stability overall.
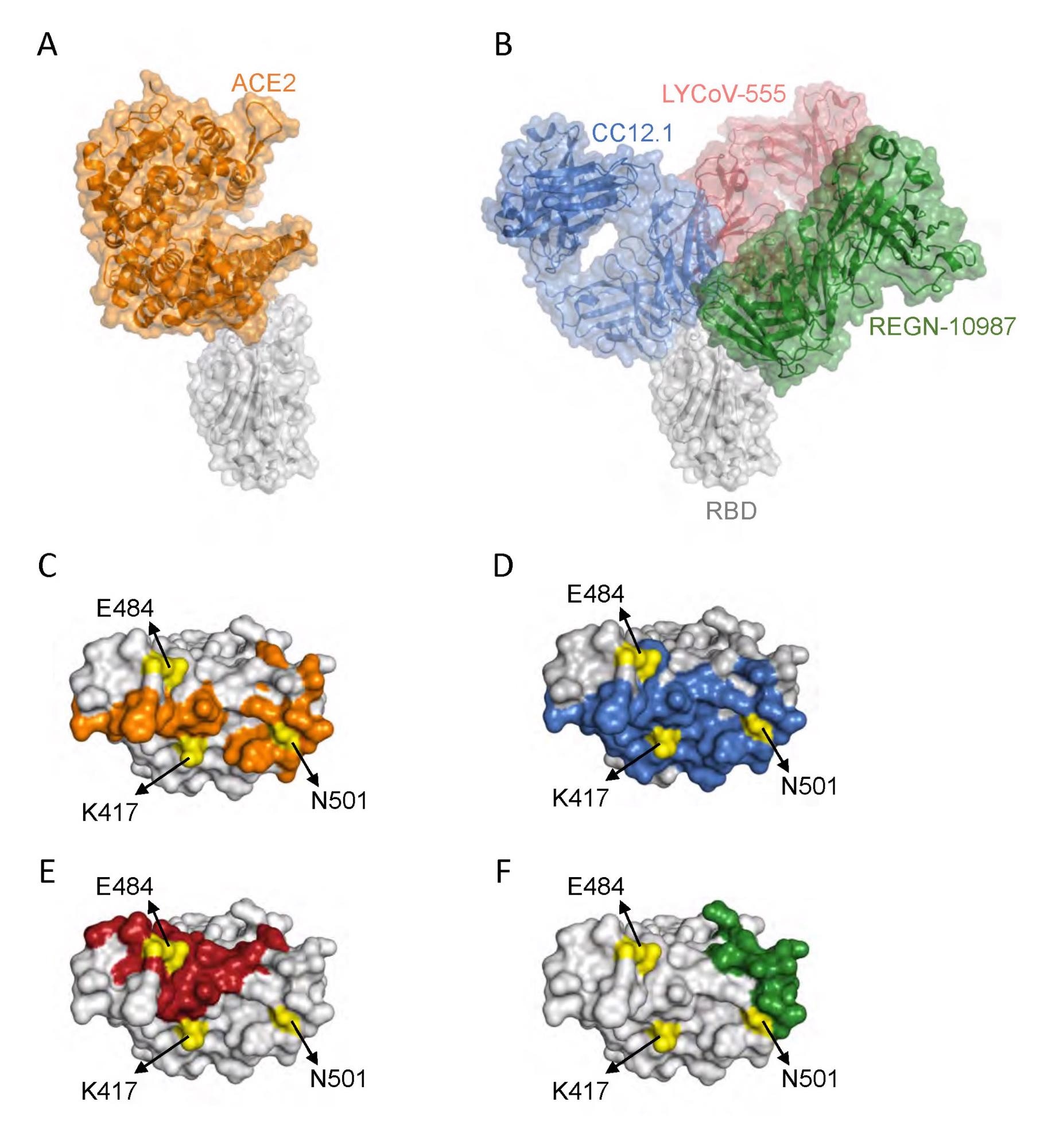 Interaction of SARS-CoV-2 RBD with ACE2 and neutralizing antibodies. (A) Protein structures showing interaction of RBD (gray) with ACE2 (orange) (Protein Data Bank (PDB) ID: 6m0j.pdb). (B) Protein structures showing interaction of RBD with a class 1 antibody Fab, CC12.1 (blue) (PDB ID: 6xc2.pdb), a class 2 antibody Fab, LYCoV-555 (red) (PDB ID: 7kmg.pdb), and a class 3 antibody Fab, REGN-10987 (green) (PDB ID: 6xdg.pdb). (C-F) The RBD residues that interact with ACE2 (orange), CC12.1 (blue), LYCoV-555 (red) and REGN-10987 (green). The position of RBD amino acid residues mutated in Gamma variant are shown in yellow.
Interaction of SARS-CoV-2 RBD with ACE2 and neutralizing antibodies. (A) Protein structures showing interaction of RBD (gray) with ACE2 (orange) (Protein Data Bank (PDB) ID: 6m0j.pdb). (B) Protein structures showing interaction of RBD with a class 1 antibody Fab, CC12.1 (blue) (PDB ID: 6xc2.pdb), a class 2 antibody Fab, LYCoV-555 (red) (PDB ID: 7kmg.pdb), and a class 3 antibody Fab, REGN-10987 (green) (PDB ID: 6xdg.pdb). (C-F) The RBD residues that interact with ACE2 (orange), CC12.1 (blue), LYCoV-555 (red) and REGN-10987 (green). The position of RBD amino acid residues mutated in Gamma variant are shown in yellow.
Following this, the researchers attempted to determine the binding affinity of the RBD mutants towards ACE2 using isothermal titration calorimetry. Wild-type showed a dissociation constant (Kd) of 10.0nM. N501Y improved this, showing a Kd of 3.4nM, while both K417T and E484K both decreased the binding affinities to 32.5nM and 51.5nm, respectively.
The scientists examined the ability of the mutants to escape from neutralizing antibodies compared to wild-type, including class I and II antibodies that bind to the RBD site and class III antibodies, which competitively bind to ACE2. K417T was found to decrease the binding affinity of the RBD to the antibody by the most significant factor, while the other two mutants showed no ability to escape. Double and triple mutants carrying K417T showed the same ability to escape. When it came to class II antibodies, K417T actually showed increased binding affinity, and N501Y showed no significant difference. E484K failed to bind the antibody. None of the mutants showed the ability to evade class III antibodies.
Conclusion
The authors suggest that K417T could provide more protein for viral assembly, providing a fitness advantage by increasing transmission and predominantly conferring the ability of the variant to escape from class I antibodies. E484K is significantly more important in escaping from class II antibodies, while N501Y improves ACE2 binding. While each of these mutations also carries potentially disadvantageous changes, all also have the potential to increase transmission and the severity of the disease. This study could help to inform drug developers and epidemiologists, and identify the exact changes the mutations confer upon the different variants.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
SARS-CoV-2 comes up with a perfect recipe for disaster: Each mutation plays a specific role in the evolution of variants of concern, Vaibhav Upadhyay, Casey Patrick, Alexandra Lucas, Krishna Mallela, bioRxiv 2021.12.23.474050; doi: https://doi.org/10.1101/2021.12.23.474050, https://www.biorxiv.org/content/10.1101/2021.12.23.474050v1
- Peer reviewed and published scientific report.
Upadhyay, Vaibhav, Casey Patrick, Alexandra Lucas, and Krishna M. G. Mallela. 2022. “Convergent Evolution of Multiple Mutations Improves the Viral Fitness of SARS-CoV-2 Variants by Balancing Positive and Negative Selection.” Biochemistry 61 (11): 963–80. https://doi.org/10.1021/acs.biochem.2c00132. https://pubs.acs.org/doi/10.1021/acs.biochem.2c00132.