
 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The SARS-CoV-2 Delta variant was previously the most dominant circulating variant in the United States and many other countries throughout the world. Since then, the SARS-CoV-2 Omicron (B.1.1.529) variant has emerged, which consists of several mutations in the spike gene and exhibits increased transmission.
Emergency-use authorized (EUA) monoclonal antibody (mAb) therapies have encountered resistance to the coronavirus disease 2019 (COVID-19) as a result of these variants of concern (VoCs). This has caused some therapies to be discontinued, thus underlining the need for new mAb candidates in development.
About the study
In the current study, researchers report the rapid identification of extremely effective SARS-CoV-2-neutralizing mAbs using high-throughput single-cell BCR sequencing from mice inoculated with pure SARS-CoV-2 RBD. To this end, they created a bispecific antibody against SARS-CoV-2 RBD using two antigen-recognition variable regions from two of the best mAb clones (Clones 2 and 6). These monospecific and bispecific antibodies bind to the RBD antigen with high affinity and avidity, effectively neutralizing historical and B.1.617 lineage viruses.
The researchers also created and characterized a humanized antibody to aid clinical development, which demonstrated great potency in vitro and in vivo against both the wild-type and Delta SARS-CoV-2 strains. The epitopes, as well as combinations of open and close RBD conformations of trimeric spike bonded to the mAbs, were seen in three-dimensional cryo-electron microscopy (EM) imaging in association with the spike-ectodomain trimer. The neutralizing efficacy of the lead mAb clones against B.1.617 variants was discovered using structure-based mutation and epitope analysis.
Study findings
Some antibodies may facilitate the spike conformational cycle to the final stage with all three RBDs open or locking the spike trimer in a pre-fusion state, thus boosting or preventing cell membrane fusion and syncytium formation, depending on the binding mode. According to the cryo-EM findings, Clone 6 Fab binds to spike trimer preferentially with at least one RBD down, effectively skewing the spike trimer towards pre-fusion states.
A down-RBD-binding Clone 6 Fab can also connect with a nearby up-RBD through a quaternary epitope found on the sidewall of the up-RBD, which interferes with ACE2 binding, effectively preventing access of the angiotensin-converting enzyme 2 (ACE2) receptor to two RBDs at the same time. When complexed with Clone 6 Fab, this bipartite binding mode is likely more stable than the single binding mode with the up-RBD alone, which explains why no spike trimer with all three RBDs open was found.
The researchers believed that the Clone 6 Fab binding to this RBD secondary epitope helps lock the spike trimer in the pre-fusion state, thus inhibiting spike-mediated cell membrane fusion by the historical virus and even the B.1.1.7 variant with increased ACE2 binding affinity. Due to the varied Clone 2 Fab binding conformations on the spike RBD, the spike trimer has been discovered in skewed states that favor RBDs up, which simulates the effect of ACE2 binding during the spike trimer’s conformational cycle.
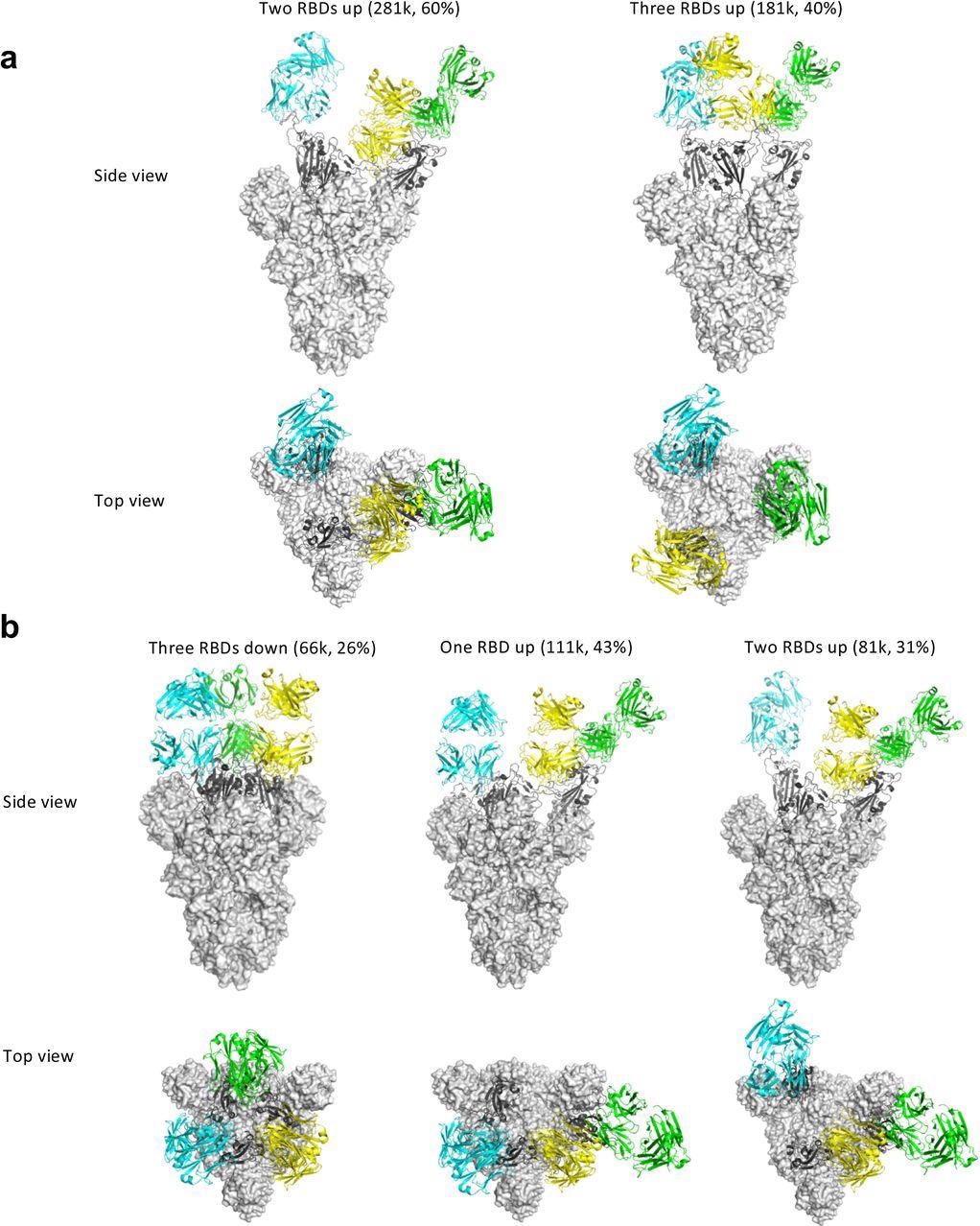
Cryo-EM structures of the ectodomain of SARS-CoV-2 spike trimer in complex with Clone 2 or Clone 6 Fab and Structural comparisons a-b, Cryo-EM structures of the ectodomain of SARS-CoV-2 spike trimer in complex with Clone 2 (a) or Clone 6 (b). Each Fab molecule is shown as ribbons in different colors, spike RBD is shown as dark gray ribbons, with the rest of spike trimer shown as gray surface. The particle distribution of each S trimer conformation is shown correspondingly.
However, the researchers discovered that Clone 2 Fab effectively reduces spike-mediated cell-cell fusion. This implies that the mechanism controlling spike-targeting antibody neutralization potency is complex.
The RBD of the B.1.1.7 SARS-CoV-2 spike features a single mutation (N501Y), which has been shown to increase the spike RBD-ACE2 binding affinity, potentially disfavoring antibody neutralizations that compete with ACE2 for RBD binding. Although N501 does not interact directly with either Clone 2 or Clone 6 Fab, it may have allosteric effects on other CDRL loops nearby, disrupting the binding interface.
In addition to the N501Y mutation, the B.1.351 variation has two additional point mutations in spike RBD, K417N, and E484K. These mutations cause both antibody clones' RBD epitope recognition to be disrupted, thereby explaining their lower potencies against the B.1.351 variant.
The researchers used the Delta RBD to perform structural analysis to better understand the influence of Delta mutations on antibody neutralizing potency. The results of this analysis showed that Clone 2 retained its neutralizing potency against the Delta variant, although REGN-10987 did not. These observations are consistent with the mutation-induced side-chain orientation, adjacent loop conformation shift, and unique antibody targeting epitopes.
Conclusion
Several prospective treatments, antibodies, and vaccines have been developed in a concerted global effort to combat COVID-19. The lead antibody clones have binding geometries and footprints that are distinct from the three antibodies described to date and are highly inhibitory against SARS-CoV-2 and various VoCs, particularly the Delta variant from the B.1.617 lineage.
“Lead antibodies showed strong inhibitory activity against historical SARS-CoV-2 and several emerging variants of concern.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Peng, L., Hu, Y., Mankowski, M., et al. (2021). Monospecific and bispecific monoclonal SARS-CoV-2 neutralizing antibodies that maintain potency against B.1.617. medRxiv. doi:10.1101/2021.12.21.473733. https://www.biorxiv.org/content/10.1101/2021.12.21.473733v1.
- Peer reviewed and published scientific report.
Peng, Lei, Yingxia Hu, Madeleine C. Mankowski, Ping Ren, Rita E. Chen, Jin Wei, Min Zhao, et al. 2022. “Monospecific and Bispecific Monoclonal SARS-CoV-2 Neutralizing Antibodies That Maintain Potency against B.1.617.” Nature Communications 13 (1): 1638. https://doi.org/10.1038/s41467-022-29288-3. https://www.nature.com/articles/s41467-022-29288-3.