Multiple Spike (S) protein mutations in the severe acute respiratory syndrome coronavirus 2 (SARS‑CoV‑2) Omicron variant contribute to the virus's escape from neutralizing antibody responses, lowering vaccination protection against infection. It's unclear to what extent other adaptive response components, such as T cells, may still target Omicron and contribute to protection against catastrophic outcomes. T cells' ability to respond to Omicron spike was tested in participants who had been vaccinated with Ad26.CoV2.S or BNT162b2, as well as in unvaccinated convalescent COVID-19 patients.
In a new study, a team of researchers from multi-national institutions discovered that 70-80% of CD4 and CD8 T cell responses to spike were consistent across research groups. Moreover, although having far more mutations, the magnitude of Omicron cross-reactive T cells was comparable to that of the Beta and Delta variants. In addition, T cell responses to ancestral spike, nucleocapsid, and membrane proteins in Omicron-infected hospitalized patients were equivalent to those seen in individuals hospitalized in earlier waves dominated by the ancestral, Beta, or Delta variations.
According to these findings, despite Omicron's widespread mutations and lower susceptibility to neutralizing antibodies, the bulk of T cell responses elicited by vaccination or spontaneous infection cross-recognize the variation. Early clinical observations from South Africa suggest that well-preserved T cell immunity to Omicron may contribute to protection from severe COVID-19.
A preprint version of this study, which is yet to undergo peer review, is available on the medRxiv* server.
Study: SARS-CoV-2 spike T cell responses induced upon vaccination or infection remain robust against Omicron. Image Credit: Fusebulb / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The study
T cell responses were studied in people who had received one or two doses of the Ad26.COV2.S vaccine (Johnson and Johnson/Janssen, n = 20 per group), two doses of the BNT162b2 mRNA vaccine (Pfizer–BioNTech, n = 15), or had recovered from infection. Convalescent donors were investigated after a median of 1.4 months of moderate or asymptomatic illness. T cell responses to vaccination were assessed 22-32 days after the final dose in more than 85% of vaccinees. Immunization and infection both produced spike-specific CD4 T cell responses, while a CD8 response was found less frequently. Intracellular cytokine labeling was used to evaluate cytokine production in response to peptide pools that covered the entire Wuhan-1 spike protein and the Omicron spike.
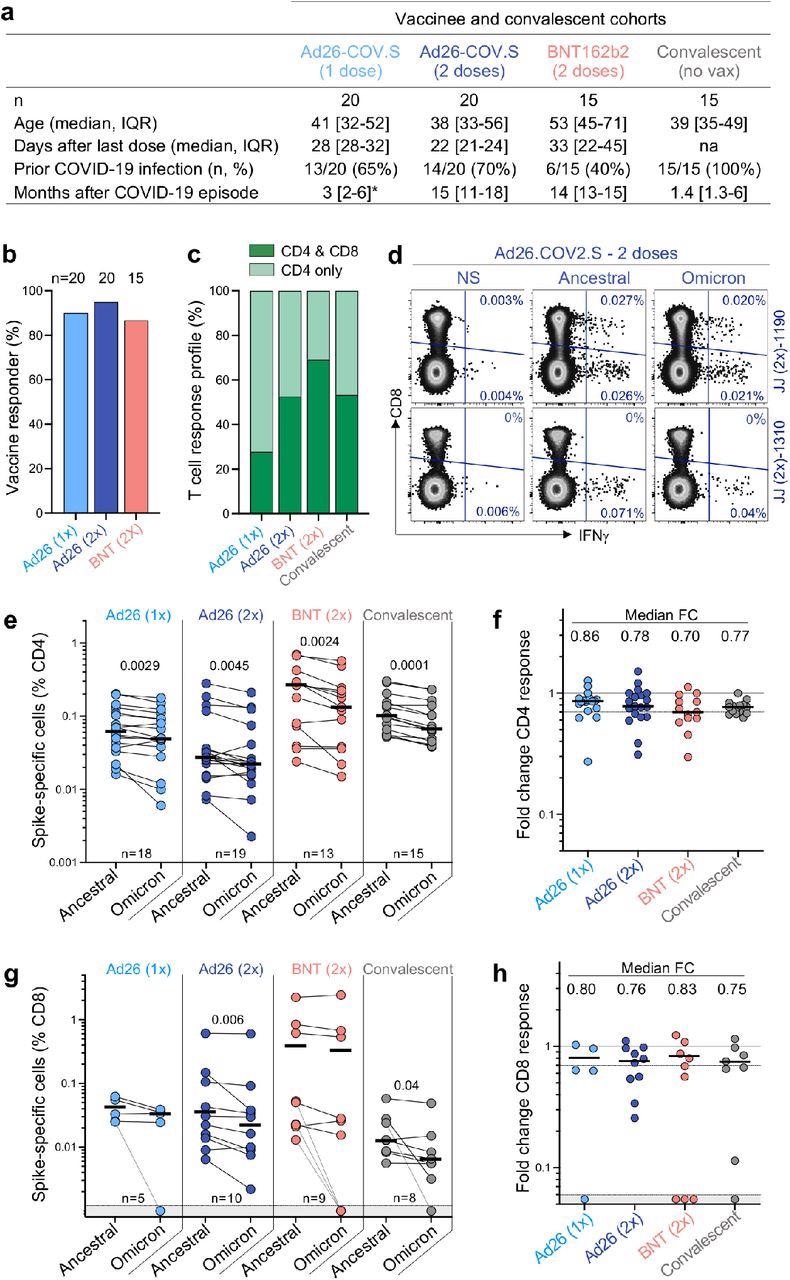
T cell response to the ancestral and Omicron SARS-CoV-2 spike after vaccination and in unvaccinated COVID-19 convalescent patients. a, Clinical characteristics of the study groups. *: data regarding time post-Covid-19 infection were available for only 6 out of the 13 participants who received one dose of Ad26.COV2.S. b, Proportion of participants exhibiting an ancestral spike-specific CD4 T cell response after vaccination with one or two doses of Ad26.COV2.S or two doses of BNT162b2. c, Profile of the ancestral spike-specific T cell response in vaccinees and convalescent individuals. d, Representative examples of IFN-γ production in response to ancestral and Omicron spike in two individuals who received two doses of Ad26.COV2.S. e,g, Frequency of spike-specific CD4 (e) and CD8 T cells (g) producing any of the measured cytokines (IFN-γ, IL-2 or TNF-α) in response to ancestral and Omicron spike peptide pools. Bars represent the median of responders. Differences between SARS-CoV-2 variants were calculated using a Wilcoxon paired t-test. f, h, Fold change in the frequency of spike-specific CD4 (f) and CD8 T cells (h) between ancestral and Omicron spike responses. Bars represent medians. No significant differences were observed between groups using a Kruskal-Wallis test with Dunn’s multiple comparisons post-test. The number of participants included in each analysis is indicated on the graphs.
In all groups studied, CD4 T cell frequencies to Omicron spike were consistently and considerably lower than the ancestral spike. This resulted in a median drop of 14-30% in CD4 responsiveness to Omicron, as evidenced by foldchange. In the case of the CD8 T cell response, vaccinees who had received two doses of Ad26.COV2.S and convalescent donors showed a significant reduction in the magnitude of Omicron spike-specific CD8 T cells, whereas the other groups did not. When compared to the ancestral virus, the CD8 response to Omicron was reduced by 17-25 %. A small percentage of responders showed a loss of CD8 T cell recognition of Omicron, which is likely due to mutations in specific CD8 epitopes affecting specific human leukocyte antigen (HLA) molecules.
T cell affinity can be reduced by mutations in variant epitopes, which can impact a cell's functional capacity. As a result, the authors evaluated the polyfunctional profiles of T cells in vaccinees and convalescent patients and found that both ancestral and Omicron-specific T cells have a similar capacity for cytokine co-expression across all groups. There were no variations in the polyfunctional profiles of CD4 or CD8 T cells between ancestral and Omicron spike, indicating that there was no functional impairment in cross-reactive Omicron T cell responses. By examining spike peptide pools matching the viral sequences of the Beta and Delta strains, the authors were able to compare Omicron spike responses to other variants of concern in Ad26.CoV2.S vaccinees.
With the exception of a higher drop in the Omicron CD4 response compared to Beta in recipients of two doses of Ad26.COV2.S, there were no significant variations in cross-reactive CD4 and CD8 T cell responses between Beta, Delta, and Omicron. Prior SARS-CoV-2 infection was linked to a larger frequency of spike-specific T lymphocytes in vaccinees but did not affect Omicron cross-reactivity. These findings reveal that Omicron spike CD4 and CD8 T cell recognition is mostly retained relative to the ancestral strain and comparable to other relevant variants with three times fewer mutations.
Implications
Overall, these findings show that vaccination and infection elicit a robust CD4 and CD8 T cell response that predominantly cross-reacts with Omicron, consistent with recent research on T cell escape by Beta, Delta, and other variants. Despite significant neutralization escape against Omicron, the T cell response is retained in 70-80% of cases. Because Omicron's mutations have a limited influence on the T cell response, immunization or prior infection may still give significant protection against severe disease.
In fact, compared to the last Delta wave, South Africa has recorded a lower likelihood of hospitalization and serious morbidity. Omicron's apparent milder effects may be due to cross-reactive T cell responses established by vaccination or infection. The T cell response's resiliency was established in this study, which bodes well for the future emergence of more heavily altered types.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
SARS-CoV-2 spike T cell responses induced upon vaccination or infection remain robust against Omicron, Roanne Keeton, Marius B. Tincho, Amkele Ngomti, Richard Baguma, Ntombi Benede, Akiko Suzuki et al, medRxiv, 2021.12.28, https://doi.org/10.1101/2021.12.26.21268380 , https://www.medrxiv.org/content/10.1101/2021.12.26.21268380v1
- Peer reviewed and published scientific report.
Keeton, Roanne, Marius B. Tincho, Amkele Ngomti, Richard Baguma, Ntombi Benede, Akiko Suzuki, Khadija Khan, et al. 2022. “T Cell Responses to SARS-CoV-2 Spike Cross-Recognize Omicron.” Nature 603 (7901): 488–92. https://doi.org/10.1038/s41586-022-04460-3. https://www.nature.com/articles/s41586-022-04460-3.