Many studies have indicated that bats are significant reservoirs of innumerable viruses, such as influenza, coronaviruses, and rabies. These studies have also reported that bats predominantly remain asymptomatic or develop mild diseases during viral infections caused by henipaviruses, Ebola virus, coronaviruses, etc.
The immune responses elicited via viral infection differs substantially between bats and human cells. Previous studies have shown that bats possess unique transcripts not present in other mammals. Bats undergo limited activation of interferon due to mutation in the STING protein and have constricted type I IFNα locus. However, they show constitutive IFNα expression without viral stimulation. The unique antiviral gene expression profile, as well as immune response pathways, make bats more tolerant against various viral infections.
It is believed that bats might be the initial host of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), the causal agent of the ongoing coronavirus disease 2019 (COVID-19) pandemic. Even though the direct progenitor of this virus has not been confirmed yet, the closest member has been identified to be the horseshoe bat (Rhinolophus sinicus). Therefore, this genus is regarded as potential reservoir hosts of SARS-CoV-2. In addition, horseshoe bats also harbor other members of coronaviruses, including SARS-CoV. This shows that horseshoe bats play a critical role in preserving coronaviruses that have the potential to cause future pandemics.
Immune responses upon viral infection are specific to species; this is why clinical symptoms in the lower respiratory tract differ among species. Although bats, cats, tigers, and pangolins are susceptible to SARS-CoV-2 infection, the biological background is not well understood.

Study: Comparative analysis of single cell lung atlas of bat, cat, tiger and pangolin. Image Credit: Binturong-tonoscarpe / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
A New Study
Scientists stated that it is vital to understand the differential immune responses induced in different animals infected by SARS-CoV-2.
Recently, a group of researchers has conducted a comparative study using single-cell transcriptomic data to determine the immune responses in the lungs of bats, cats, tigers, and pangolins. This study has been posted to the bioRxiv* preprint server while awaiting peer review.
This study's primary objective is to elucidate the molecular basis of differential immune responses in the studied animals. Researchers collected lung tissues from healthy bats to create single-nucleus libraries of lung cells, and a total of 11,838 pulmonary cells passed quality control.
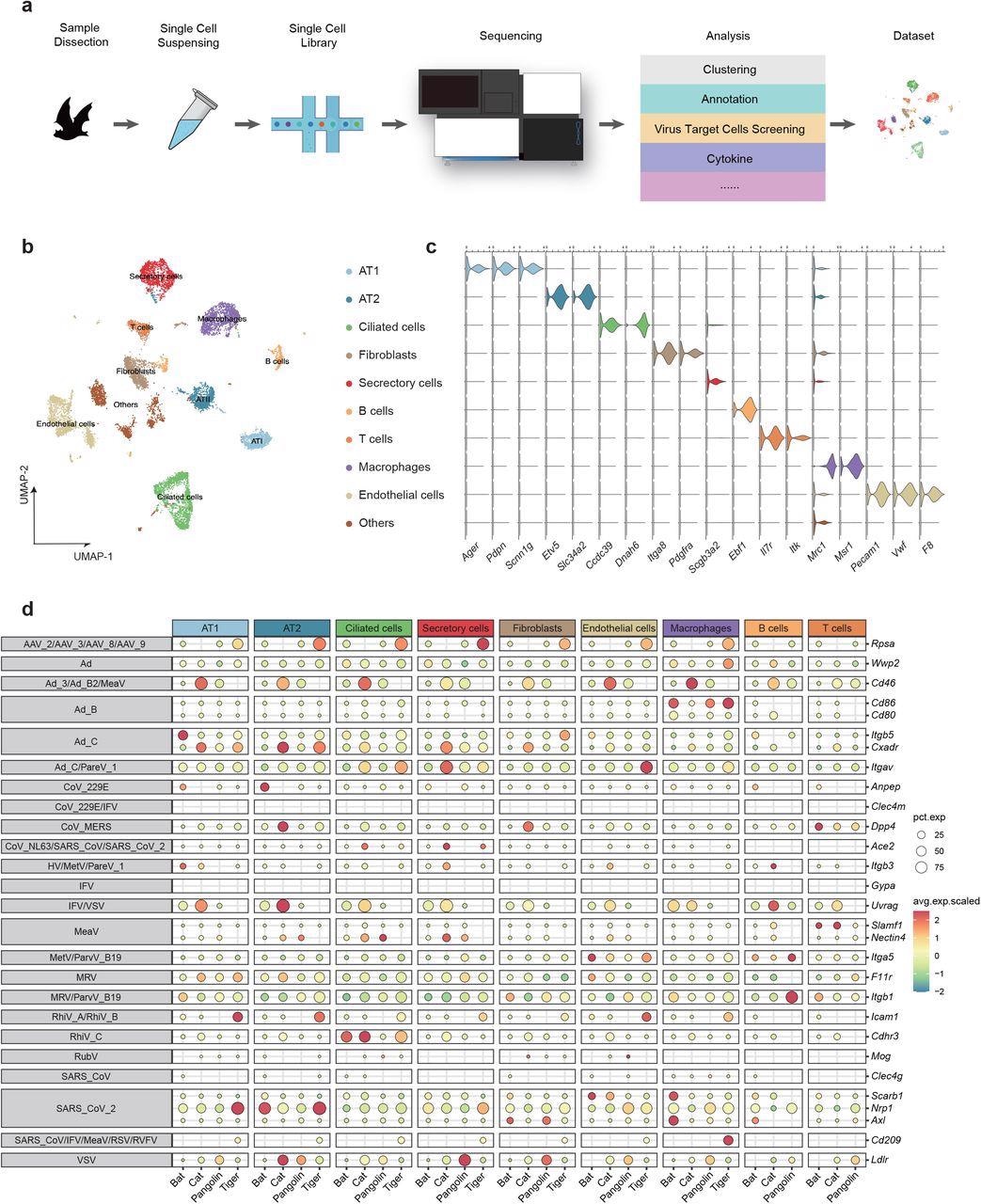
Comparative single-cell lung atlas of the bat, cat, tiger and pangolin. a, Illustration of the overall project design. b, UMAP plot of bat lung. c. Violin plot showing the expression patterns of canonical cell type markers. d. Expression proportion and scaled expression value of virus receptors in distinct cell types of bat, cat, tiger and pangolin.
Main Findings
Researchers identified nine major cell types in the lungs of bats that include ciliated cells, alveolar type 1 cells (AT1), alveolar type 2 cells (AT2), secretory cells, endothelial cells, fibroblasts, B cells, T cells, and macrophages. Each of these cells was recognized via specific cell type markers.
Additionally, scientists studied the expression patterns of twenty-nine genes that encode entry receptors of respiratory viruses in the lung cells of the four studied species. This is important because receptor binding domains are crucial for viral infection and the distribution of the receptors determines the susceptibility of cells to a particular viral infection. Additionally, when a virus binds to these receptors, it induces the immune response in the host cell.
The scientists found that, compared to pangolins, tigers, and cats, Itgb5, a receptor of adenoviruses, was abundantly present in bat AT1 and Anpep, a receptor of human coronavirus 229E, was enriched in bat AT2. However, Cd86, also an adenovirus receptor, was abundantly found in the macrophages of bats, pangolins, and tigers. Cdhr3, a rhinovirus receptor, was primarily found in the ciliated cells of both bat and cat. Rpsa is another adeno-associated virus receptor, was found to be significantly expressed in tiger pulmonary cells except in T and B cells.
SARS-CoV and SARS-CoV-2 infect host cells using ACE2 receptor, which was abundantly expressed in ciliated and secretory cells of the cat. ACE2 receptor was reported to be expressed in the secretory cells of a tiger. However, only marginal ACE2 expression was found in bat endothelial cells and macrophages. Scarb1 is another receptor for SARS-CoV-2, which exhibited highly specific expression in endothelial cells and macrophages of bats. Two other SARS-CoV-2 receptors, namely, Nrp1 and Axl, showed high species specificity. Nrp1 was reported to be expressed in the AT1/AT2 of tigers and AT2 of bats. Axl was found to be highly expressed in fibroblasts and macrophages of the bat lung and fibroblasts of the pangolin lung.
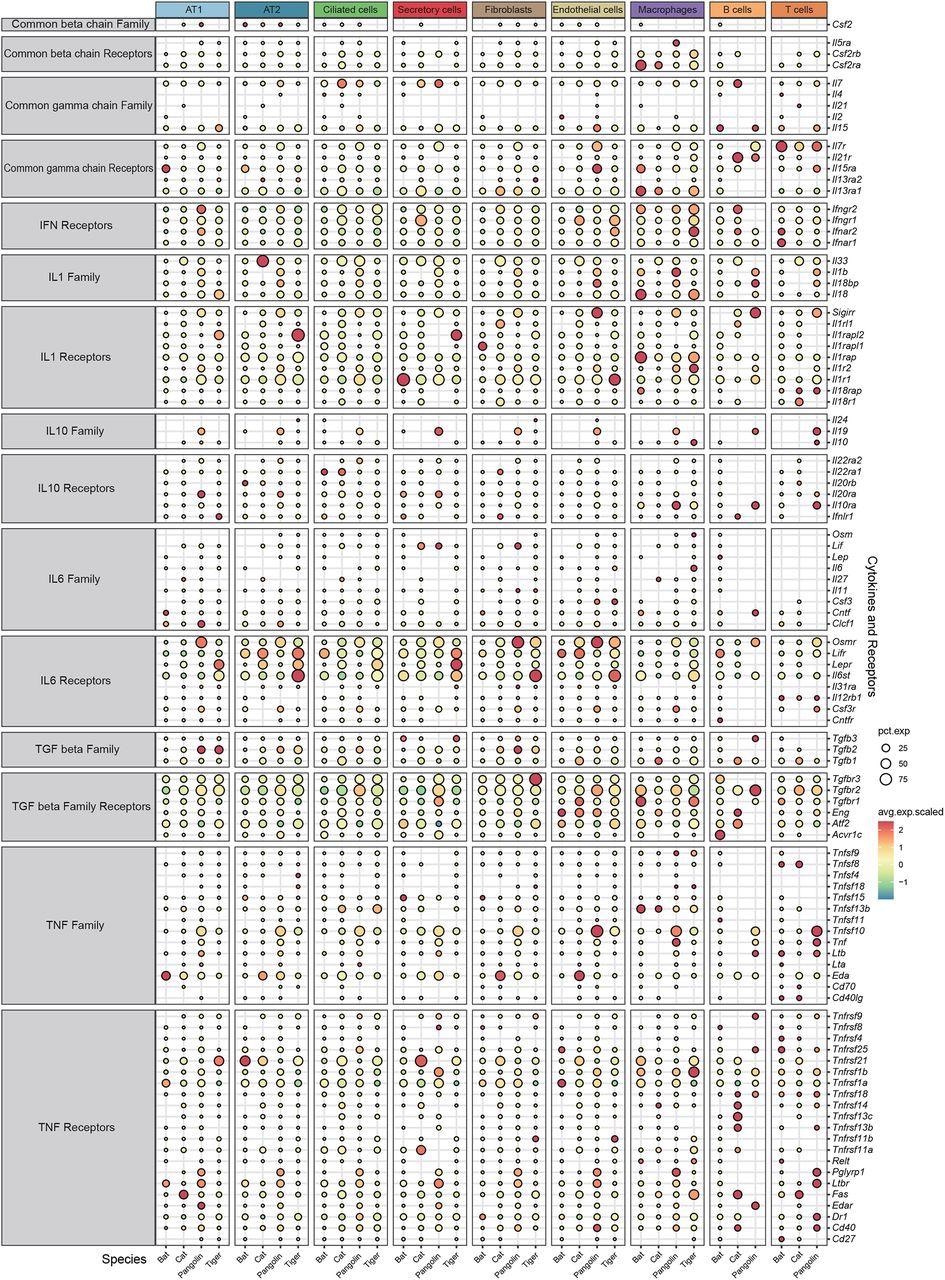
Interestingly, bats do not display any significant symptoms after virus infection, such as cytokine storm, one of the main contributing factors of severe lung pathogenesis caused by various virus infections, including SARS-CoV-2. Scientists did not find any significant differences in the anti-inflammation cytokines (IL10, TGF beta) and corresponding receptors among the pulmonary cells of the four studied species. However, expression of receptors for specific pro-inflammation cytokines (IL1, IL6, TNF, interferons), IL-6 (Osmr, Lifr), and interferons (IFN) were considerably low in bat pulmonary cells. Interestingly, two receptors of type 1 interferon (Ifnar1, Ifnar2) and a receptor for IFN-γ (Ifngr2) were found to be depleted in bat cells.
Conclusion
The authors revealed that the transcriptions of some pro-inflammatory cytokine receptors were suppressed in bat lung cells. This finding presents a novel insight into the specific immune characteristics of bats. However, controlled infection experiments are required to validate the results of this study.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Wang, X. et al. (2022) Comparative analysis of single cell lung atlas of bat, cat, tiger and pangolin, bioRxiv, 2021.12.26.473325; doi: https://doi.org/10.1101/2021.12.26.473325, https://www.biorxiv.org/content/10.1101/2021.12.26.473325v1
- Peer reviewed and published scientific report.
Wang, Xiran, Peiwen Ding, Chengcheng Sun, Daxi Wang, Jiacheng Zhu, Wendi Wu, Yanan Wei, et al. 2022. “Comparative Analysis of Single Cell Lung Atlas of Bat, Cat, Tiger, and Pangolin.” Cell Biology and Toxicology, September. https://doi.org/10.1007/s10565-022-09771-9. https://link.springer.com/article/10.1007/s10565-022-09771-9.