On November 26, 2021, the SARS-CoV-2 Omicron (B.1.1.529) variant of concern (VOC) was first reported in South Africa. As compared to the previously dominant Delta variant, preliminary analyses show that Omicron has a higher transmission rate, higher immune evasion potential, and lower virulence.
Study: From Delta to Omicron: analyzing the SARS-CoV-2 epidemic in France using variant-specific screening tests (September 1 to December 18, 2021). Image Credit: PHOTOCREO Michal Bednarek / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
About the study
In the current study, three mutations in the SARS-CoV-2 spike protein were studied from the 131,478 screening tests. These mutations included E484K, E484Q, and L452R, all of which were referred to as mutation A, mutation B, and mutation C, respectively. The researchers used A0B0C1 to refer to Delta variant infections, A0B0C0 to refer to Alpha or Omicron variant infections or an ancestral lineage, and A1B0C0 to refer to Beta or Gamma variant infections.
The researchers utilized a multinomial regression model to determine characteristics related to the result of variant-specific screening test between October 25, 2021, and December 18, 2021, when the pandemic was on the rise.
A0B0C0 infections were more common in young people than A0B0C1 infections. In most of the French regions with high relative risk ratios (RRR), the researchers also found significant temporal increases.
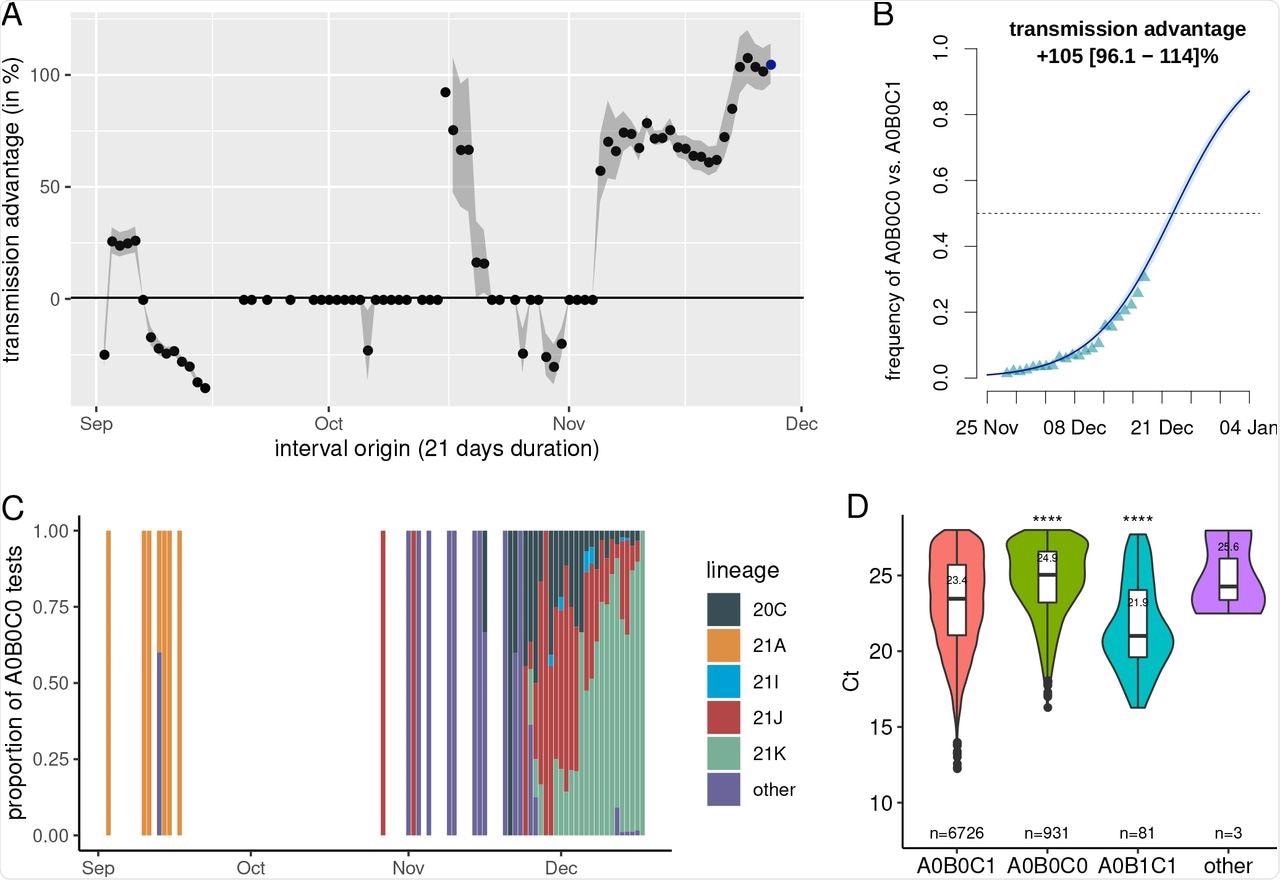
A) Transmission advantage of A0B0C0 tests over A0B0C1 in France, B) Estimated frequency of A0B0C0 relative to A0B0C0 and A0B0C1 tests in France, C) Lineage of the infected associated with A0B0C0 tests using full genome sequencing (N = 1,610), and D) Cycle threshold values as a function of the test results (N = 7,741).
Next, the researchers calculated the transmission advantages of A0B0C0 infections over A0B0C1 infections during a 21-day period. Because variant screening techniques only looked for three mutations, the researchers sequenced the entire genome of 1,160 A0B0C0 samples.
The researchers used a linear model to compare the cycle threshold (Ct) values with age, administrative region, sampling site, and sample date as the primary determinants. To reduce bias in screening test findings only tests with a Ct value less than or equal to 28 were included. The sample size was the sole factor that did not show up in an analysis of variance (ANOVA) with a type II error.
The researchers investigated two scenarios for the beginning of 2022 using the inferred transmission advantage by updating a previous epidemiological model customized to the French epidemic, which has been shown to yield reliable findings over a five-week horizon. The model is based on two types of immunity including population natural immunity, which is a model output, and vaccination immunity, which is based on national data.
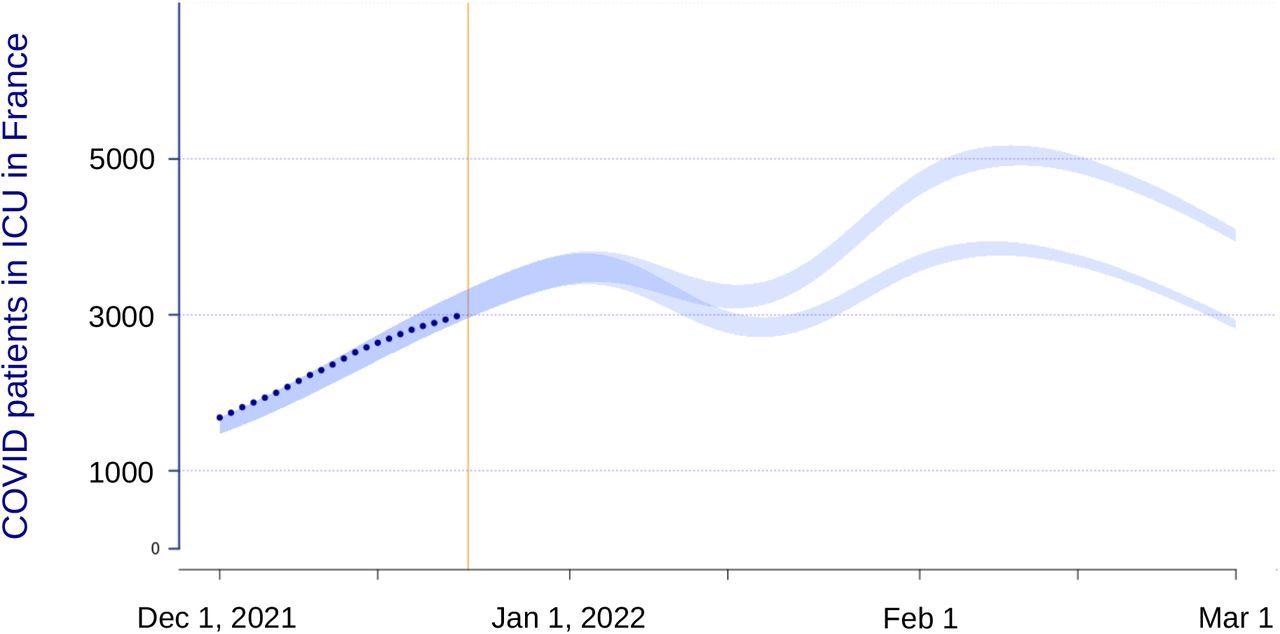
National intensive care unit dynamics in two scenarios of Omicron properties. The vertical yellow line indicates the day the model was performed, the dark blue dot the data, and the shaded envelope the compatibility intervals of the model projections.
Study takeaways
The study findings described a rapid rise of Omicron variant infections in France, which is consistent with what has been observed in other nations around the world. The researchers demonstrate that an increasing fraction of A0B0C0 tests corresponds to the spread of the Omicron variant by combining variant-specific screening tests on all positive samples with full-genome sequencing on some of the samples.
The Ct values of A0B0C0 samples were substantially higher than those of A0B0C1. Although caution should be exercised when interpreting Ct values, particularly for coronaviruses, this indicates a lesser amount of virus genetic material in the samples, which is consistent with early findings of changes in tissue tropism between the Omicron and Delta variants.
The fact that the Omicron variant has a reduced virus burden in nasopharyngeal swabs has serious implications for reverse-transcription polymerase-chain-reaction (RT-PCR) test sensitivity.
Lastly, epidemiological modeling demonstrated that even though the virulence of the Omicron variant is lower than that of the Delta variant, the rise in reproduction number from the data has the ability to keep coronavirus disease 2019 (COVID-19) activity high in French hospitals, without overburdening them. As a result, rapid response to the latest epidemic wave is critical.
“A0B0C0 samples exhibited significantly higher cycle threshold (Ct) values than A0B0C1 samples.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Sofonea, M. T., Roquebert, B., Foulongne, V., et al. (2022). From Delta to Omicron: analyzing the SARS-CoV-2 epidemic in France using variant-specific screening tests (September 1 to December 18, 2021). medRxiv. doi:10.1101/2021.12.31.21268583. https://www.medrxiv.org/content/10.1101/2021.12.31.21268583v1.
- Peer reviewed and published scientific report.
Sofonea, Mircea T., Bénédicte Roquebert, Vincent Foulongne, David Morquin, Laura Verdurme, Sabine Trombert-Paolantoni, Mathilde Roussel, et al. 2022. “Analyzing and Modeling the Spread of SARS-CoV-2 Omicron Lineages BA.1 and BA.2, France, September 2021–February 2022.” Emerging Infectious Diseases 28 (7): 1355–65. https://doi.org/10.3201/eid2807.220033. https://wwwnc.cdc.gov/eid/article/28/7/22-0033_article.