
 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Early reports from South Africa have shown that the SARS-CoV-2 Omicron variant has higher transmissibility and immune evading capability than the previously dominant Delta variant. In addition, evidence from contact tracing and household infection studies has prompted speculation that Omicron generation time may be shorter than the generation time of the Delta variant in similar settings.
The authors of the current study have previously estimated growth rates for the Omicron and Delta variants jointly in the UKHSA region. They demonstrated that the growth advantage of the Omicron variant versus the Delta variant appears to vary over time.
About the study
In the present study, the researchers estimated the generation time distribution and transmission advantage of the Omicron variant that would be required to explain the observed time-varying growth advantage of Omicron. While the researchers assumed a gamma-distributed generation distribution for Omicron generation time estimates, they repeated this estimation process using two different prior forecasts for the generation time of the Delta variant. The first estimation was based on household transmission, whereas the other was based on the intrinsic generation time of the Delta variant.
The researchers examined the data from November 23, 2021, to December 23, 2021, the transition period during which the Omicron variant replaced the Delta variant as the dominant circulating strain. During this time period, people were generally not seeking coronavirus disease 2019 (COVID-19) tests due to the holidays.
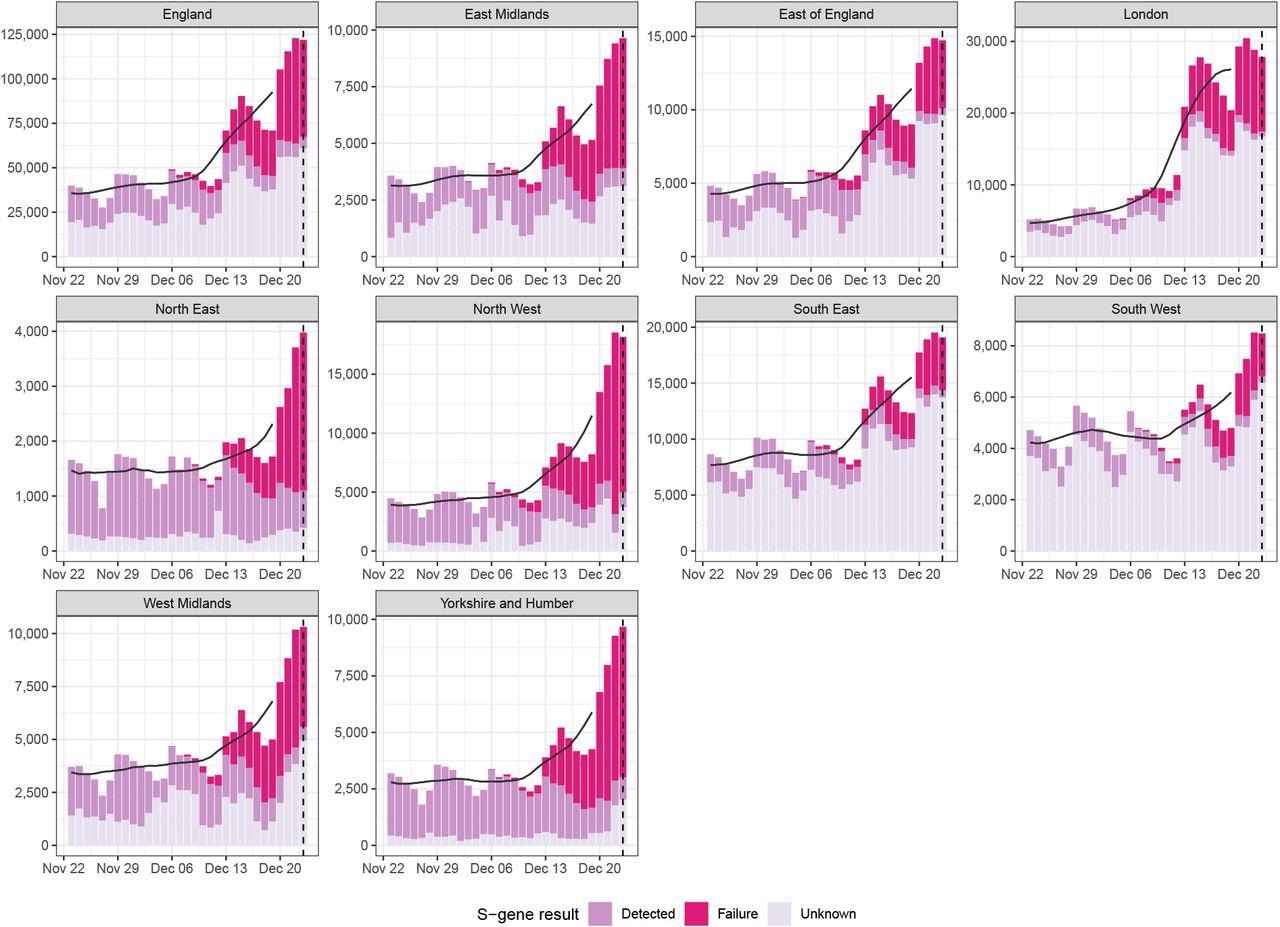
Daily cases in England and by UKHSA region, with S-gene target result (failed, confirmed detected, or unknown), and centred 7-day moving average starting from the 23rd of November 2021 and including data up to the 23rd of December 2021 (dotted line). Source: UKHSA and coronavirus.gov.uk; data by specimen date.
S-gene target failure (SGTF), a marker for the Omicron variant, showed the proportion of Omicron-positive test samples among SARS-CoV-2 positive samples. The researchers augmented this data with total reported surveillance case counts. Both data sets were sourced from the UKHSA region by date of specimen and SGTF status. All the data was assessed using an autoregressive (AR) model that allowed for estimating time-varying differences in the transmission advantage of the Omicron variant.
Following an AR approach allowed the researchers to model expected cases for each variant as a combination of expected cases from the previous day and the exponential log growth rate for that variant and time point. This model accounted for variant differences other than a transmission advantage. In the absence of evidence that variants do not co-vary, it was reduced to a model with a fixed transmission advantage.
The gamma approximation framework modeled a relationship between estimated variant growth rates, generation times, and the variant transmission advantage. Using this approximation, the effective reproduction number (RD) of the Delta variant was estimated across locations (l) and times (t, in days) using estimates of the daily growth rates (rD) and a gamma-distributed generation time. The gamma approximation framework was used in reverse to estimate the daily growth rates for Omicron (rO) and compare these results to previous estimations.
Study findings
The initial evidence for a difference in generation time distributions was provided by visualizing the growth rate estimates of the study.
The assumed generation time distribution for the Delta variant was a mean of 2.5-4 days and a standard deviation of 1.9-3 days. With a mean of 1.5-3.2 days and a standard deviation of 1.3-4.6 days, the estimated generation time distribution for Omicron was shorter than the Delta variant.
The findings from this study demonstrate that the Omicron variant has a transmission advantage of 160%-210% as compared to the Delta variant. The estimates of the intrinsic generation time for Delta were comparable, although those estimates increased in magnitude due to the longer assumed generation time.
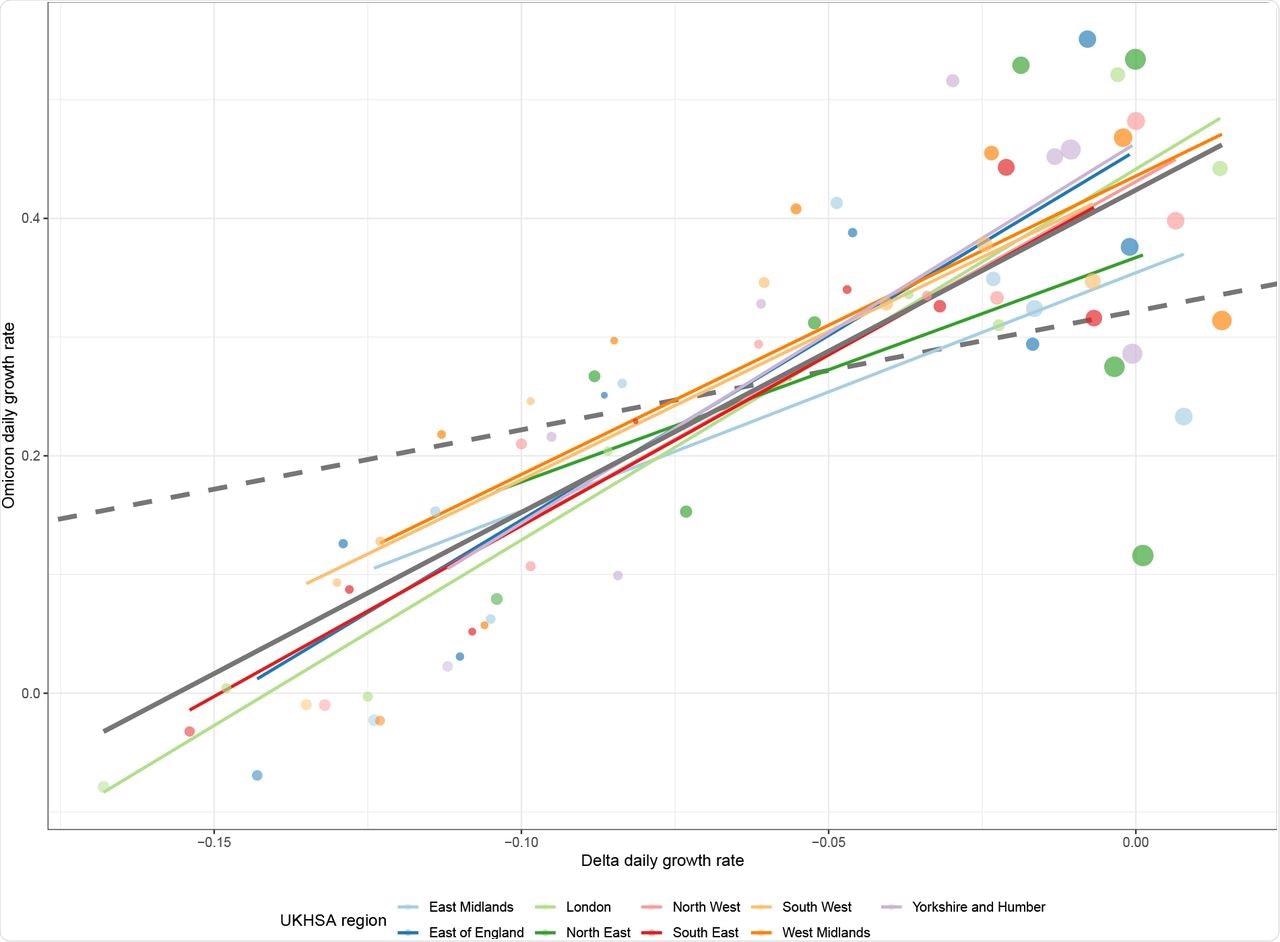
Estimates of the time-varying daily growth rate for each UKHSA region for cases with the Omicron variant and non-Omicron cases. Point size has been used to represent the uncertainty of each estimate (though it does not correspond to a meaningful credible interval). The dashed grey line indicates the expected relationship between variants if they differ only by a fixed transmission advantage. The solid grey line indicates the observed overall linear relationship between variants, and the slope of this line approximates the ratio between generation time means. The region-specific lines show how this relationship differs between regions.
Conclusions
To summarize, a reduction in the generation time of Omicron compared to Delta explained the observed variation over time in the transmission advantage of the Omicron variant.
The role of other factors could not be ruled out, including the impact of differential immune escape by variants and sampling bias in the SGTF data. Similarly, differences in age group among populations in which variants are circulating and the varying generation times between variants could have also impacted the observed variations.
The authors recommend further exploration of these additional factors using similar methods to those described in this study. The model described in the study could potentially be extended to a latent infection-based model or stratified by age. Alternative study designs, such as household transmission studies and contact tracing studies, could also be used for such explorations.

 *Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: medRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.