Coronaviruses (CoVs) are single-stranded, positive-sense Ribonucleic Acid (RNA) viruses that belong to the Coronaviridae family's Orthocoronavirinae subfamily. Alphacoronavirus (α-CoV), Betacoronavirus (β-CoV), Gammacoronavirus (ϒ-CoV), and Deltacoronavirus (δ-CoV) are the four genera in which they are categorised.
Coronaviruses are significant human and animal pathogens that cause a variety of diseases. Deltacoronaviruses and gammacoronaviruses primarily infect birds with some mammalian spillover, whereas alphacoronaviruses and betacoronaviruses primarily infect mammals.
During virological monitoring in southern China between 2005 and 2006, deltacoronaviruses were found in the rectal swabs of mammalian species in wet markets, including Asian leopard cats and Chinese ferret badgers.
In Hong Kong, the prototype porcine deltacoronavirus (PDCoV) HKU15 strain was identified in 2012. However, it was not discovered to have pathogenic potential until 2014, when it was found to be the source of pig diarrhea epidemics, which began on numerous farms in Ohio and then spread across the United States (US) and around the world.
In both gnotobiotic (living in germ-free conditions) and conventional pigs, PDCoV infects intestinal epithelial cells, causing severe watery diarrhea, vomiting, and dehydration. Other swine enteric CoVs, such as porcine epidemic diarrhea virus (PEDV) and transmissible gastroenteritis (TGEV) virus, cause clinical symptoms similar to those caused by PDCoV.
The present state of knowledge about the genesis, evolution, cross-species transmission, and zoonotic potential of PDCoV is the subject of a review published in the journal Pathogens, which was conducted in a combined effort by researchers from Heilongjiang Bayi Agricultural University and Ohio State University.

Study: Porcine Deltacoronaviruses: Origin, Evolution, Cross-Species Transmission and Zoonotic Potential. Image Credit: SGr / Shutterstock
The origin of PDCoV
The International Committee on Taxonomy of Viruses (ICTV) first recognized the Deltacoronavirus genus in 2012, following the identification of three novel avian coronaviruses (bulbul CoV HKU11, thrush CoV HKU12, and munia CoV HKU13) [6] and seven other novel δ-CoVs (porcine CoV HKU15, white-eye CoV HKU16, sparrow CoV HKU17, magpie robin CoV HKU18, night heron CoV HKU19, wigeon CoV HKU20, and common moorhen CoV HKU21) of birds or pigs. The replicase polyprotein gene of PDCoV HKU15 is interestingly connected to the deltacoronavirus discovered in 2006 in Chinese ferret badgers at wet markets. HKU15's polyprotein, spike (S) protein, envelop (E) protein, membrane (M) protein, non-structure protein 6 (NS6), nucleocapsid (N) protein, and non-structure protein 7 (NS7) genes had 98.70% nucleotide identity with those of the δ-CoV identified in an Asian leopard cat.
More notably, a recent retrospective investigation in Anhui Province, China, showed the prevalence of PDCoV in diarrheic pigs as early as 2004.
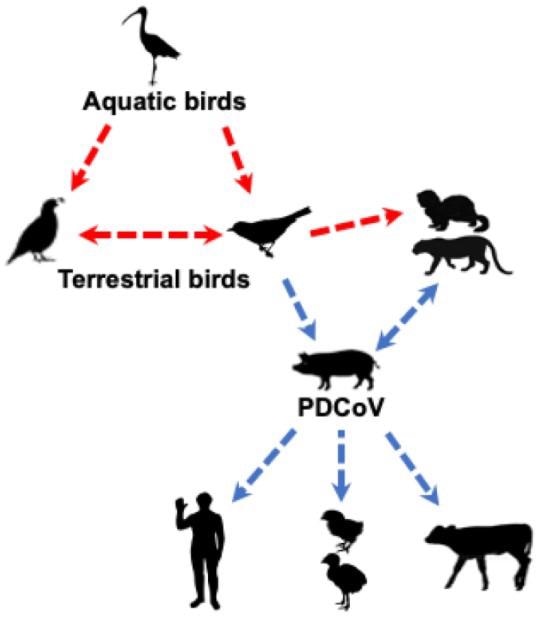
Likely origin and routes of cross-species transmission of PDCoV. The red dashed line indicates potential, but unknown, transmission of δ-CoVs from avian to mammalian species; the blue dashed line indicates potential transmission of PDCoV based on epidemiology or experimental studies.
Molecular epidemiology and diversity of PDCoVs
PDCoV was initially discovered in mainland China in 2015, but historical investigations reveal that it could have been a swine disease as early as 2004. Ninety-four of 719 swine diarrhea samples collected from 18 provinces in China between March 2016 and June 2018 were PDCoV-positive by RT-qPCR, according to a recent PDCoV surveillance study. The S genes of 11 PDCoV strains were determined in their entirety. The Chinese lineage was assigned to seven people. CH-WH-2017, on the other hand, was more closely related to the US/Japan/South Korea lineage. PDCoV has been found in 26 Chinese provinces, including all swine-producing areas, as of October 2021.
PDCoV-related diarrhea epidemics were discovered in Canada, South Korea, Thailand, Vietnam, and Laos from mid-March 2014 to January 2016. In addition, PDCoV has been circulating in Taiwan, Japan, Mexico, and Haiti since 2011, late 2013, 2015, and 2014, according to retrospective investigations. Although retrospective studies have been undertaken in Brazil and Spain to find CoVs in wild birds and pigs, no PDCoVs or PDCoV-like-CoVs have been detected in Europe, South America, Africa, or Australia.
The zoonotic potential of PDCoVs
Three zoonotic CoVs (SARS-CoV, MERS-CoV, and SARS-CoV-2) have posed a severe threat to public health and global security in the previous two decades. Also of zoonotic origin are HCoV-HKU1, HCoV-229E, HCoV-NL63, HCoV-OC43, and HECV-4408. A complete genome of a novel canine-feline recombinant alphacoronavirus (CCoV-HuPn-2018, GenBank accession no. MW591993.2) isolated from a child with pneumonia in Malaysia was first characterized in a recent study. Following this discovery, another investigation discovered and described a canine-feline recombinant alphacoronavirus in a medical team member that traveled to Haiti.
In November 2021, a study published the first indication that the PDCoV virus had spread from pigs to three Haitian children. The three children only had minor symptoms and were able to recover. These findings imply that PDCoV was transmitted from pigs to humans in at least two separate instances. PDCoV has only been shown to produce asymptomatic or moderate illness in humans, and it does not appear to pose a significant health concern to humans. In the future, however, continuous surveillance and seroprevalence investigations of PDCoV in humans, particularly those in direct contact with pigs, may provide certainty.
Conclusion
To further understand the ecology and development of deltacoronaviruses, more research into their genetic diversity in avian and mammalian hosts is required. Experiments on PDCoV interspecies transmission and host adaptation are also needed to assess the virus's pathogenicity in different hosts.
Furthermore, seroprevalence investigations in pigs can provide valuable information about the global prevalence of PDCoV. Finally, to determine the impact of PDCoV on public health, ongoing surveillance and seroprevalence investigations in people are required. Pigs may require safe and efficient vaccines to prevent infection and to diminish the PDCoV gene pool, which contributes to spillovers, and humans may require vaccines if severe cases occur.
Journal reference:
- Porcine Deltacoronaviruses: Origin, Evolution, Cross-Species Transmission and Zoonotic Potential, Fanzhi Kong, Qiuhong Wang, Scott P. Kenney, Kwonil Jung, Anastasia N. Vlasova, and Linda J. Saif, MDPI, 2022.01.09, https://doi.org/10.3390/pathogens11010079, https://www.mdpi.com/2076-0817/11/1/79/htm