A recent report published in the United States Centers for Disease Control and Prevention (CDC) Emerging Infectious Diseases explores the dynamics of viral interference between various respiratory viruses in light of the ongoing coronavirus disease 2019 (COVID-19) pandemic.
Virus-virus interactions occur when different respiratory viruses concurrently infect the respiratory tract and subsequently lead to either a synergistic or antagonistic effect on replication or infection of the viruses. Many studies have evaluated the mechanisms of viral interferences in animal models susceptible to the respiratory viruses of interest and differentiated airway epithelial cells. One such mechanism is interferon (IFN)-mediated temporary non-specific immunity in the host.
An increase in disease severity or pathogenesis of viruses is associated with a positive virus-virus interaction. Negative virus interactions depend upon whether the viruses belong to the same or different family.
In the case of homologous virus interactions, cross-reactivity from the first virus inhibits the growth of the second. In heterologous interactions, the induction of non-specific innate immunity by the first virus prevents or reduces the infection associated with the second virus.
Viral interference
In the 1960s, a research group described the concept of viral interference by developing live enterovirus vaccines (LEVs) to prevent various enteric diseases, such as polioviruses. Apart from the protection these vaccines offer against pathogenic enteroviruses, oral administration of LEVs in children lowered the detection of several unrelated viruses such as human adenovirus, influenza virus, and parainfluenza, which may be due to viral interference caused by LEV-induced IFN.
Similarly, a study conducted between 1968–1971 demonstrated possible LEV-associated protection against acute viral respiratory infections. However, these findings received backlash due to some rare incidences of vaccine-associated paralytic poliomyelitis and vaccine-derived poliovirus during that period.
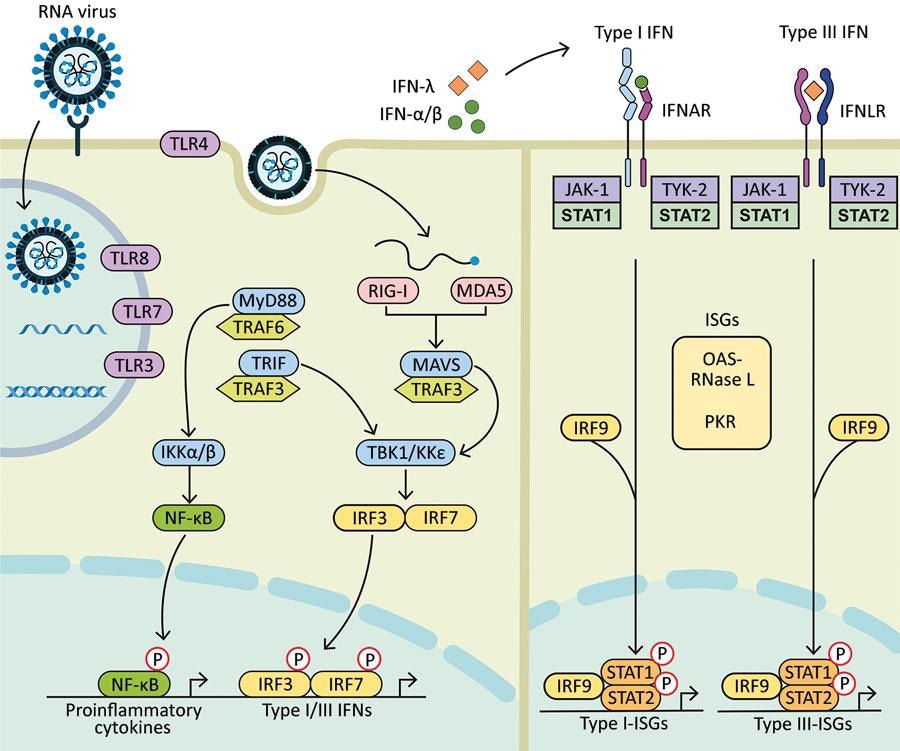
Diagram showing how components of RNA viruses are recognized by TLRs located at the plasma membrane (TLR4, viral glycoprotein sensing) and in the endosomal compartment (TLR3, double-stranded RNA sensing; TLR7 and TLR8, both single-stranded RNA sensing). Virus replication intermediates and replicated genomes are also recognized by cytosolic RNA sensors, RIG-I, and MDA5. Downstream adaptor proteins, MyD88 for TLR4, TLR7, and TLR8; TRIF for TLR3 and TLR4, and MAVS (for MDA5 and RIG-I) are activated. These activations trigger signaling cascades through TRAF3 and TRAF6; TBK1; and IKKα, IKKβ, and IKKε, which leads to phosphorylation and nuclear translocation of NF-κB, IRF3, and IRF7. These changes result in the production of proinflammatory cytokines and type I and type III IFNs. Secreted IFN-α/β and IFN-λ bind to their specific receptors (IFNAR and IFNLR) in infected and neighboring cells. Activation of JAK-1 and TYK-2 leads to phosphorylation of STAT1 and STAT2. After translocation in the nucleus, phosphorylated STAT1 and STAT2 form a complex with IRF9 to induce expression of ISGs, such as OAS-RNase L and PKR, and establishment of an antiviral program. IFN, interferon; IFNAR, IFN-α/β receptor; IFNLR, interferon-λ receptor; IKK, an inhibitor of nuclear factor-κB kinase; ISGs, IFN-stimulated genes; IRF, IFN regulatory factor; JAK-1, Janus kinase 1; MAVS, mitochondrial antiviral signaling protein; MDA5, melanoma differentiation-associated gene 5; MyD88, myeloid differentiation factor 88; NF-κB, nuclear factor-κB; OAS, 2′-5′ oligoadenylate synthetase; P, phosphorylated protein; PKR, protein kinase receptor; RNase L, latent endoribonuclease; RIG-I, a retinoic acid-inducible gene I; STAT, signal transducer and activator of transcription; TBK 1, TANK binding kinase 1; TLRs, Toll-like receptors; TRAF, tumor necrosis factor receptor-associated factor; TRIF, TIR-domain-containing adapter-inducing IFN-β; TYK-2, tyrosine kinase 2.
Interferences between respiratory viruses
Several epidemiologic studies have demonstrated potential viral interferences at the host level.
The types and subtypes of influenza virus include influenza A(H1N1), seasonal influenza A(H3N2), and influenza A(H1N1)pdm09 pandemic virus (pH1N1). During the sequential and mixed outbreak of the influenza virus in Japan in the 1977–78 winter season, it was observed that antibody generation in the sequential and viral interference in the mixed outbreaks was responsible for the cross-subtype protection.
One study conducted in ferrets investigating the potential viral interference between influenza virus types pH1N1 and influenza B virus (IBV) subtypes pH1N1 and H3N2 indicated a viral interference when the primary and secondary infections occur within three to seven days. Further, pH1N1 virus infection in ferrets prevented a subsequent infection with the respiratory syncytial virus (RSV) for about seven days.
Following a primary RSV infection, low morbidity was associated with secondary pH1N1 viral infection, though all ferrets were co-infected. Although pH1N1 viral infection generated more chemokines, cytokines, and immune mediators as compared to RSV in the respiratory tract, both infections generated a low amount of cross-reactive IFN-γ-producing cells. These findings suggested an innate immunity-based interference between IAV and RSV.
The differentiated human lung epithelial cells pre-infected with RSV reduced the likelihood of a subsequent human metapneumovirus (HMPV) infection, whereas the reverse effect was not observed. This viral interference was associated with IFN-α, IFN-λ, IFN-I, and IFN-III.
The differentiated human airway epithelial cells pre-infected with human rhinovirus (HRV) conferred significant protection against successive pH1N1 or IAV infections for about three days by inducing interferon-stimulated gene (ISG) and inhibiting IFN signaling pathways with BX795. In mouse models, HRV was less potent in preventing IAV infection concomitantly; however, two days before the HRV inoculation, lowered morbidity and mortality was associated with IAV through an early controlled pulmonary inflammatory response.
Children with prior exposure to RSV had a lower incidence of HRV than those without a previous RSV infection. However, those who were co-infected showed an increase in disease severity as compared to a single RSV infection. Furthermore, infants who received palivizumab for immunoprophylaxis against RSV had a higher incidence of HRV as compared to those without this medication history.
Influenza virus, HRV, and SARS-CoV-2
Although the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) generates a broader range of ISGs, chemokines, and cytokines in the human nasal mucosa than the pH1N1 virus, an early IFN-I and IFN-III response in human lung tissues were not associated with SARS-CoV-2 as compared to the influenza virus. Thus, the virus-virus interaction depends upon the initial virus causing the infection.
Studies with golden Syrian hamsters have indicated that an initial pH1N1 infection reduced the lower pulmonary SARS-CoV-2 load if the SARS-CoV-2 infection occurs after a day. However, prior SARS-CoV-2 infection did not affect the pH1N1 load in the lungs as compared to a single pH1N1 infection. Co-infection of SARS-CoV-2 and pH1N1 viruses led to an increase in disease severity and lung inflammatory damage when compared to the effect of single infections.
One limitation of this study was the insufficient time between viral infections to develop an interference. During the influenza viral infection in ferrets, a non-specific innate immunity was elicited by them after two to three days, during which the second virus was inoculated also. Hence, more studies are required to understand SARS-CoV-2 and influenza virus interactions.
Despite the mitigation measures implemented during the COVID-19 pandemic, HRV remained prevalent throughout the period. Since HRV is a nonenveloped virus, it is resistant to sanitizing, and the use of face masks does not prevent transmission of HRV.
Studies have demonstrated that human bronchial epithelial cells previously infected with HRV impaired SARS-CoV-2 replication by inducing several ISGs, whereas the opposite is not true. However, BX795-mediated inhibition of ISG induction will overcome HRV interference and lead to a higher SARS-CoV-2 replication rate.
Defective viral genomes
Defective viral genomes (DVGs) synthesized during the replication of ribonucleic acid (RNA) viruses play a significant role in viral adaptation, host immune escape, and persistence. DVGs can inhibit cytopathic effects produced by a wild-type virus.
Moreover, by competing with viral genomes for packaging or replication, DVGs mediate homologous viral interference and, through the production of IFN-I and IFN-III, heterologous interference. Hence, DVGs have been used as therapeutic agents against respiratory virus infections.
Conclusions
Once the sanitary restrictions implemented during the COVID-19 pandemic are lifted, other respiratory virus interactions with SARS-CoV-2 can be estimated. In the case of closely spaced co-exposures of viruses sharing similar ecological conditions, the initial viral infection might induce a refractory period. During this time, there are lower chances of subsequent respiratory virus infection in the host.
Factors influencing viral interferences include the rapid induction of IFN by the interfering virus, the extent of susceptibility of the second virus towards immune mediators and different viruses' ability to counteract IFN induction, as well as the degree of innate immunity activated in the respiratory tract by each virus. Viral interference at the population level is difficult to demonstrate since there are differences in the transmission dynamics of the respiratory viruses according to social behaviors and the time of year among various age groups.
Prospective epidemiologic studies can demonstrate viral interference by assessing multiple respiratory viruses in serial nasopharyngeal swab specimens of subjects from various epidemic periods. The interaction between respiratory viruses causing different epidemic peaks should be evaluated for their chances of co-detection and mechanisms using in vivo and ex vivo models.