The current coronavirus disease 2019 (COVID-19) pandemic has led to the development of more than 200 vaccine candidates; however, only a few of these vaccines have been licensed for human use. Most of the current COVID-19 vaccines encode the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) full-length spike (S) protein, including two stabilizing proline mutations (S2P).
Although vaccines against SARS-CoV-2 have been developed at an unprecedented speed, questions regarding the efficacy and durability of protective immunity remain, primarily due to the emergence of several SARS-CoV-2 variants of concern (VOCs).

Study: An antibody targeting the N-terminal domain of SARS-CoV-2 disrupts the spike trimer. Image Credit: Preechar Bowonkitwanchai / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
Many groups have studied antibodies against VOCs, focusing on potency and structure. It has also been reported that unrelated individuals can produce genetically and functionally similar clones of antibodies post-vaccination or infection.
The receptor-binding domain (RBD) of the SARS-CoV-2 S protein interacts with the human angiotensin-converting enzyme 2 (ACE2) receptor to gain entry inside cells. Additionally, the N-terminal domain (NTD) of S also interacts with receptors or co-receptors to promote viral attachment and infection via the ACE2 receptor pathway.
The NTD also helps to evade neutralization by binding biliverdin through the recruitment of tetrapyrrole rings. The NTD can also exhibit conformational flexibility that results in wide tissue tropism. Therefore, the functional qualities of the human antibody response against NTD are not completely understood.
A new study published on the preprint server bioRxiv* discusses the targeted discovery of NTD-reactive antibodies from an individual who had recovered from SARS-CoV-2 infection with the help of the single-B-cell barcoding LIBRA-seq antibody discovery technology.
About the study
The current study involved an investigation of peripheral blood B-cells from four individuals who had a history of laboratory-confirmed symptomatic SARS-CoV-2 infection, as well as one healthy donor who served as a negative control. Antigen-specific B-cell sorting was performed using cells that were stained and mixed with DNA-barcoded antigens and other antibodies. Thereafter, sample preparation, library preparation, and sequencing of the DNA-barcoded antigens were carried out followed by bioinformatic analysis.
High-throughput antibody expression was performed with the help of microscale transfections followed by monoclonal antibody (mAb) production and purification. The researchers also involved several assays including enzyme-linked immunosorbent (ELISA) binding assays, cell-surface antigen-display assay, focus reduction neutralization test (FRNT), high-throughput real-time cell analysis (RTCA) neutralization assay, and conventional throughput neutralization assay.
Serum antibody competition binding ELISAs were performed using biotinylated reference mAbs. Animal studies using female heterozygous K18-hACE C57BL/6J mice were carried out to determine the protection against SARS-CoV-2 in mice. Finally, epitope mapping of antibodies was carried out by alanine-scanning mutagenesis followed by measurement of viral burden using plaque assays.
Study findings
The results reported that the four SARS-CoV-2 positive individuals showed the highest reactivity against the SARS-CoV-2 S2Pecto, RBD, and NTD proteins. Moreover, the serum antibody reactivity of one individual was found to be highest against the NTD protein, as well as high serum neutralizing titers. Neutralizing titers were found to remain high, even after three months from infection.
Most antibodies were found to recognize NTDs, with one individual possessing 347 NTD-specific B-cells. Nine mAbs with full or partial neutralizing capacity were also identified.
The IGHV1-24 and IGHV1-69 variable gene segments of antibodies were found to be most frequently associated with an individual's response. Out of these antibodies, the most potent neutralizing NTD-reactive antibodies were encoded by IGHV1-24.
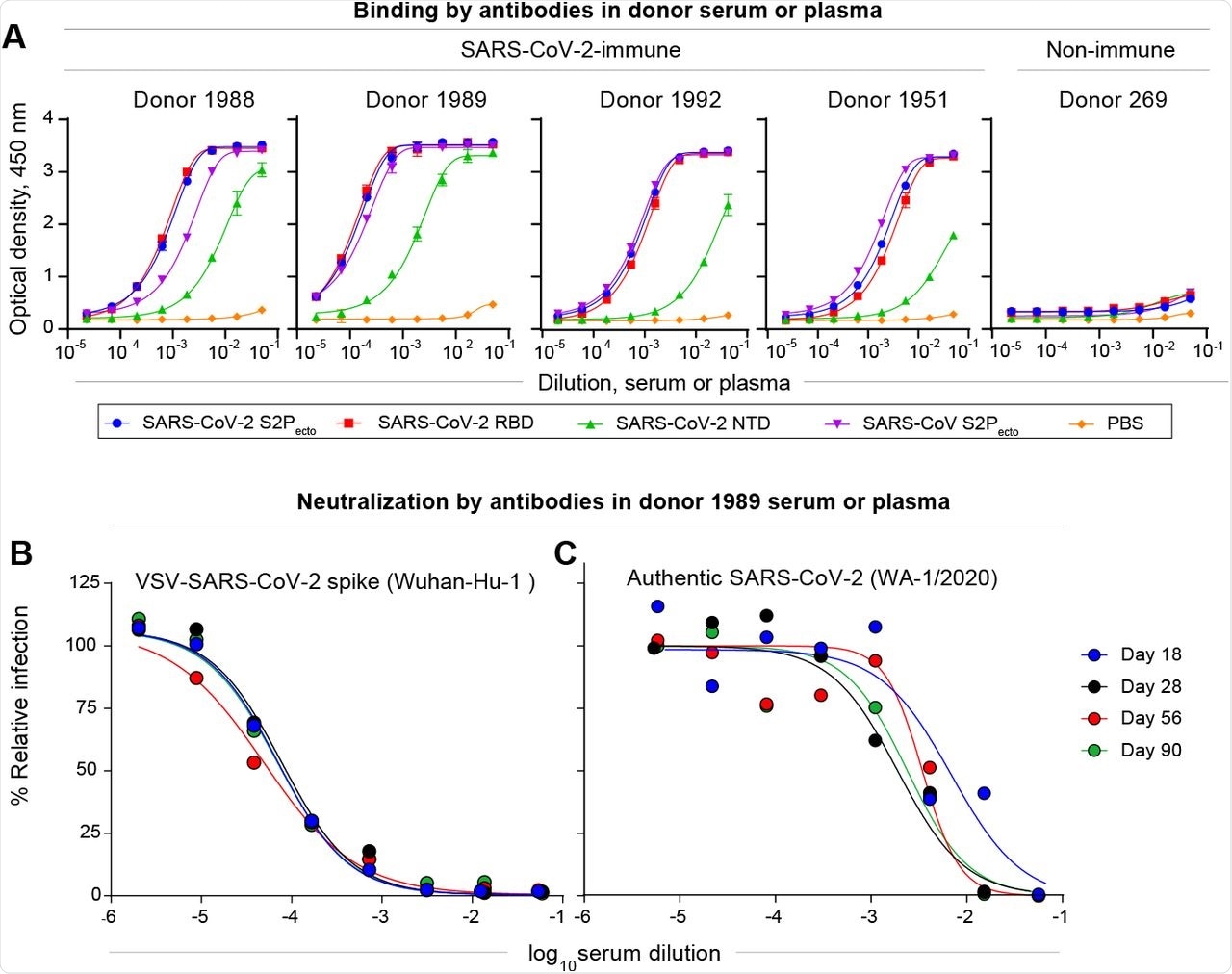
Characterization of SARS-CoV-2 antibodies in convalescent patient samples.
The results also indicated that the COV2-3434 mAb binding site on NTD is unique and did not compete with other identified mAbs to bind NTD. The binding of COV2-3434 mAb was found to be associated with disruption of the S protein trimer of SARS-CoV-2. Moreover, the binding of COV2-3434 was found to take place only when the RBD adopts an ‘up’ state conformation of the S6Pecto protein.
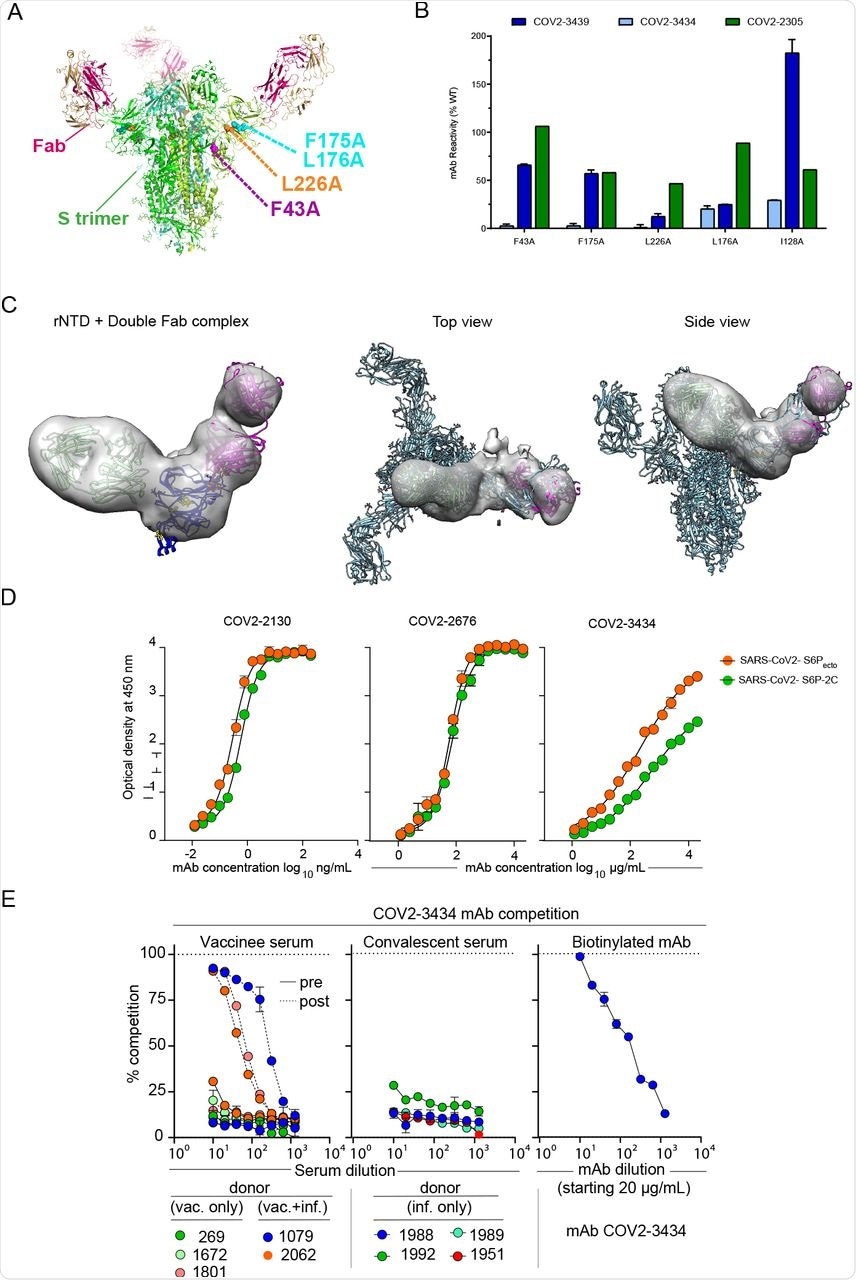
Structural characterization of the trimer-disrupting antibody COV2-3434.
Furthermore, up to 90% serum antibody competition with COV2-3434 was reported in three SARS-CoV-2 positive individuals post-vaccination, while no such competition was reported in the case of convalescent individuals. The COV2-3434 mAb was also found to provide protection against VOCs, as well as K18-hACE2-transgenic mice that were challenged with SARS-CoV-2.
Conclusions
Taken together, the current study demonstrates that COV2-3434 is a rare SARS-CoV-2 S trimer interface antibody that plays an important role in the neutralization of SARS-CoV-2. These antibodies may also be found in the serum of several vaccinated individuals.
Therefore, knowledge of the impact of these antibodies on vaccine protection is important especially due to the emergence of VOCs. In addition, these antibodies can play an important role in the updated vaccine designs.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Suryadevara, N., Shiakolas, A. R., VanBlargan, L. A., et al. (2022). An antibody targeting the N-terminal domain of SARS-CoV-2 disrupts the spike trimer. bioRxiv. doi:10.1101/2022.01.12.476120. https://www.biorxiv.org/content/10.1101/2022.01.12.476120v1.
- Peer reviewed and published scientific report.
Suryadevara, Naveenchandra, Andrea R. Shiakolas, Laura A. VanBlargan, Elad Binshtein, Rita E. Chen, James Brett Case, Kevin J. Kramer, et al. 2022. “An Antibody Targeting the N-Terminal Domain of SARS-CoV-2 Disrupts the Spike Trimer.” The Journal of Clinical Investigation 132 (11). https://doi.org/10.1172/JCI159062. https://www.jci.org/articles/view/159062.