SARS-CoV-2 is the causative agent of the coronavirus disease 2019 (COVID-19) pandemic that has caused an unprecedented public health crisis leading to a global state of emergency. It is an RNA virus with a linear, positive-sense, single-stranded genome approximately 30 Kb-long encoding 29 proteins. SARS-CoV-2 has four conserved structural proteins - nucleocapsid protein (N), spike protein (S), an envelope protein (E), and membrane glycoprotein (M).
SARS-CoV-2 variants and their characteristics
In the present review, the authors discussed the characteristics of SARS-CoV-2 and its variants covering various aspects, including preventive measures with an emphasis on physical and material science-based approaches.
The S protein has been extensively studied because it has significant implications in the infectivity of SARS-CoV-2. It consists of two subunits - S1 and S2; the S1 subunit harbors the N-terminal domain (NTD) and receptor-binding domain (RBD). The S protein mediates the entry of virions in the host cells via the angiotensin-converting enzyme-2 (ACE2) by facilitating the fusion of host and viral membranes.
![A) Illustration of S protein-ACE2 binding, and location of the frequently detected mutations within the spike region of SARS-CoV-2. B) Detailed visualization of mutations that are found within the genomic structure of current VoC lineages. To display amino acid changes within the S proteins, genomic sequence data of each variant (OU030649.1, OU092214.1, OU061529.1, MZ357460.1, and OL672836.1) was obtained from GenBank database, National Center for Biotechnology Information.[66-71] Comparative 3D illustrations of the S proteins for each variant were made using CoVsurver, with mutation analysis of hCov-19 tool provided by GISAID using the hCoV-19/Wuhan/WIV04/2019 as the reference strain.[72] Adapted from “The SARS-CoV-2 Variants of Concern,” by BioRender.com (2021). Created with BioRender.com.](https://www.news-medical.net/images/news/ImageForNews_702085_16426486167184678.jpg)
A) Illustration of S protein-ACE2 binding, and location of the frequently detected mutations within the spike region of SARS-CoV-2. B) Detailed visualization of mutations that are found within the genomic structure of current VoC lineages. To display amino acid changes within the S proteins, genomic sequence data of each variant (OU030649.1, OU092214.1, OU061529.1, MZ357460.1, and OL672836.1) was obtained from GenBank database, National Center for Biotechnology Information.[66-71] Comparative 3D illustrations of the S proteins for each variant were made using CoVsurver, with mutation analysis of hCov-19 tool provided by GISAID using the hCoV-19/Wuhan/WIV04/2019 as the reference strain.[72] Adapted from “The SARS-CoV-2 Variants of Concern,” by BioRender.com (2021). Created with BioRender.com.
Variants of SARS-CoV-2 have been classified into categories: variant of concern (VOC) and a variant of interest (VOI) or variant under investigation (VUI).
In November 2020, the first VOC named Alpha (B.1.1.7) was detected in the United Kingdom (UK). It was associated with high transmission rates and increased disease severity, causing more deaths than the wild-type SARS-CoV-2. In addition, a mutation, N501Y, was noted in its S protein that increased its affinity to ACE2, thereby increasing the potency of infectivity.
Sometime later, another VOC was identified in South Africa and was classified as a Beta variant (B.1.351) which contained the E484K substitution within the S RBD and the K417N and L18F mutations in the S protein resulting in subpar immune responses.
SARS-CoV-2 P.1 VOC was the third variant that emerged in Japan and Brazil in early 2021 with a highly mutated S. Reports identified many mutations and found that 12 were within the RBD at key sites similar to previous variants.
The Delta variant, B.1.617, initially observed in India in April 2021, rapidly spread and replaced the circulating variants. It had notable mutations such as L452R, T478K, and P681R associated with high transmission and immune escape features.
The SARS-CoV-2 Omicron (B.1.1.529) variant is the latest VOC detected in November 2021 in South Africa. Compared with the original virus, Omicron has 30 amino acid changes, three small deletions, and one small insertion in the spike protein. In the bronchi (lung airways), Omicron multiplies about 70 times faster than the Delta variant. Still, it appears to be less severe than previous strains, especially when compared to the Delta variant. It may be harder for Omicron to penetrate deep lung tissue. Compared to the delta variant, Omicron infections result in 91 percent fewer deaths and 51 percent less hospitalization. Due to its extremely high rate of spread and its ability to evade both double vaccinations and the body's immune system, the number of patients requiring hospital treatment at any given time remains very high. Many mutations have been identified in this variant, some of which have concerned scientists. The Omicron variant has a total of 60 mutations compared to the reference/ancestor variant: 50 nonsynonymous mutations, 8 synonymous mutations, and 2 non-coding mutations. Twenty-two mutations affect the spike protein, the main antigenic target of antibodies that are generated during infections and by many vaccines. Many of those mutations have not been previously identified in previous strains.
Vaccines and drugs against COVID-19
Vaccination is the most effective and efficient strategy to control infectious diseases. With over 9.7 billion doses of COVID-19 vaccines administered globally, they have significantly contributed to preventing the spread of the virus, decreasing hospitalizations and disease severity, and lowering the impact of the pandemic.
Countries with high vaccine resources have achieved higher vaccination rates than low-resource nations, and this inequity in vaccine access and distribution can create a breeding ground for more variants threatening regions with robust vaccination rates.
Antiviral therapeutics such as Remdesivir inhibits RNA replication of SARS-CoV-2 and is the most commonly used antiviral for COVID-19 treatment. The use of antibiotics has also been explored in the context of COVID-19 treatment. The glycopeptide antibiotic teicoplanin inhibits cathepsin L, which is vital for viral entry into cells. Monoclonal antibody and convalescent plasma therapies have also been suggested for COVID-19 treatment as additional measures.
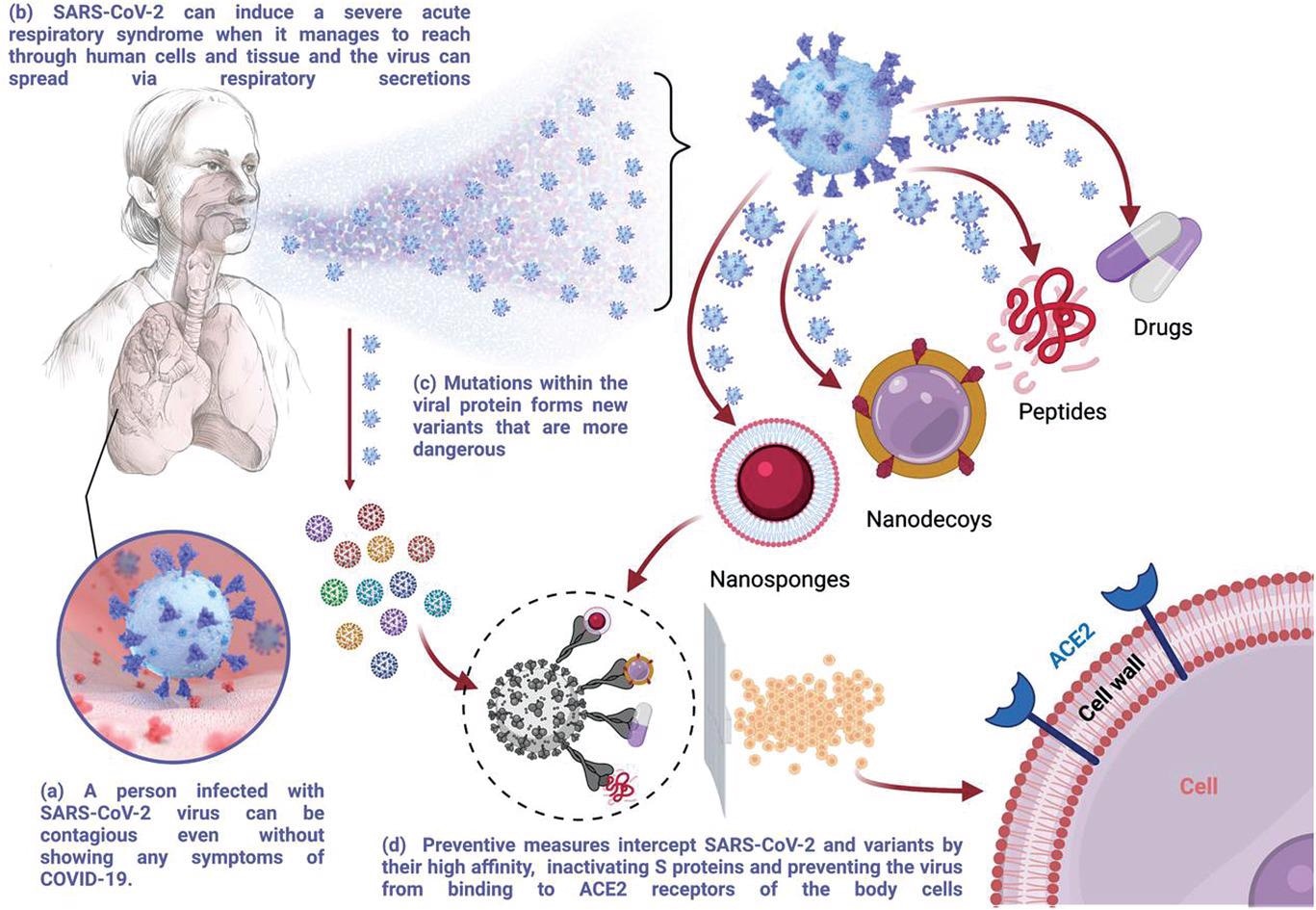
Schematic illustration of the SARS-CoV-2 transmission and scope of preventive measures discussed in this review. Created with BioRender.com.
Some novel therapies at the molecular level have been designed based on the attachment of the S protein to ACE2. Studies examined the infectivity of SARS-CoV-2 by deploying decoy targets for therapy. For instance, a study conducted on high ACE2-expressing human embryonic kidney (HEK)-293T cells with membrane-based nanoparticles enriched with ACE2 revealed that SARS-CoV-2 S selectively interacted with ACE2-nanoparticles which neutralizes the binding of S to the actual ACE2 expressed by the HEK cells, leaving them uninfected.
Inorganic polyphosphate (polyP), which is naturally secreted from platelets, has been shown to disrupt S attachment to host ACE2 in a nano-encapsulated form in another study. In another novel development, a polymeric composite (gellan and λ-carrageenan) is being evaluated in nasal form for prophylaxis and prevention of COVID-19. Furthermore, a green-tea ingredient, epigallocatechin gallate, has been shown to prevent SARS-CoV-2 infection before cell entry.
Material science approaches to developing virus-repellant surfaces can effectively prevent indirect viral transmission by contact. The positively charged SARS-CoV-2 membrane can be repelled by creating positively charged surfaces based on the fundamental concept of electrostatic repulsion. Surfaces created by electrospraying with positively charged chitosan were shown to repel viruses that could be tested for SARS-CoV-2.
In conclusion, the present review explored various possible interventions like clinical therapeutics, physical science- and material science-based methods to counter COVID-19. While more research is required, physical and material science-based approaches could be promising because these interventions are independent of viral evolution and mutations.