
 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
Over the past two years, SARS-CoV-2 has evolved by garnering numerous mutations in the receptor-binding domain (RBD) of its spike protein. The SARS-CoV-2 RBD binds to the angiotensin-converting enzyme 2 (ACE2) receptor, which allows the virus to invade host cells and replicate.
The RBD is also the region that is targeted by neutralizing antibodies induced by either natural infection or vaccination. The amino acid substitutions of the SARS-CoV-2 RBD have been reportedly associated with increased transmissibility, reduced plasma neutralization, and, most notably, immune escape characteristics. The ability of SARS-CoV-2 variants to escape immunity is alarming, particularly given the fact that more than eight billion doses of COVID-19 vaccines have been administered around the world to date.
Before the implementation of COVID-19 vaccines, monoclonal antibody (mAb) therapies were the primary tools to treat patients with severe COVID-19 and continue to be used even after the availability of vaccines. However, the selection of mAbs becomes challenging as a result of the increased number of RBD mutations and, as a result, the varying effectiveness of mAbs against different variants. Therefore, it is essential to quickly identify SARS-CoV-2 variants in clinical specimens to optimize the mAb therapies against specific variants of SARS-CoV-2.
About the study
In the present study, researchers developed an assay to quickly identify functional mutations in the SARS-CoV-2 spike protein. Rapid and accurate genotyping of critical mutations can be made possible by a droplet digital polymerase chain reaction (ddPCR). Reverse transcriptase ddPCR (RT-ddPCR) was carried out by the researchers on extracted total nucleic acid samples.
ddPCR occurs within oil-separated droplets, followed by a TaqMan probe detection in which amplification causes the separation of dye on the 5'end of the probe that is bound to the template from the quencher on its 3' end releasing the fluorescent dye. Poor interaction or binding between the probe and template due to sequence differences leads to less frequent cleavage of the dye, thereby resulting in poor or low fluorescence inside the droplet.
The fluorescence is measured after the PCR, in which the amplitude or brightness of fluorescence inside the droplet represents the affinity of binding between the probe and template. This assay, therefore, allows the mutational state of the template to be ascertained.
Study findings
In the current study, the authors tested 419 clinical specimens collected between February 2021 and December 2021. These samples were marked by the prevalence of various SARS-CoV-2 variants and underwent three duplex RT-ddPCR reactions, each of which targeted four amino acids.
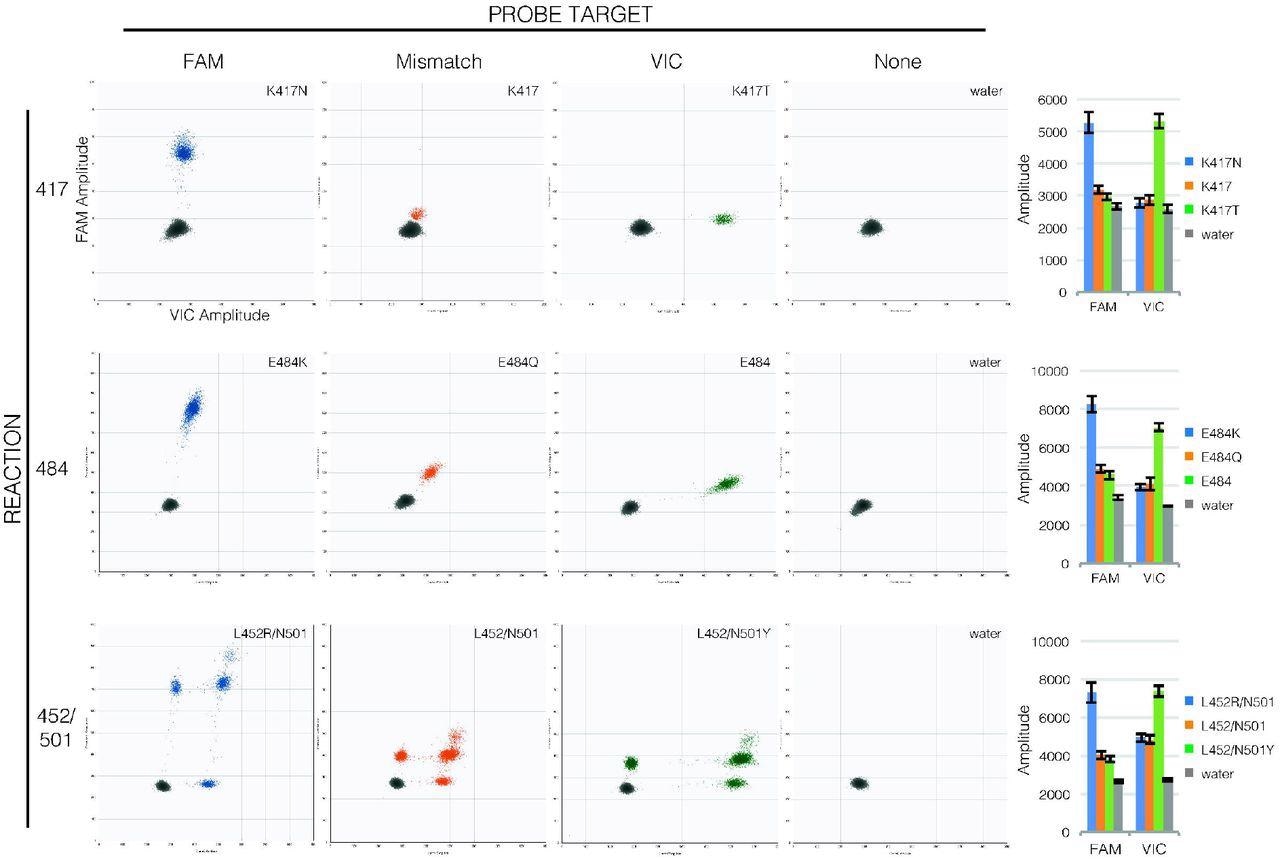
Probe binding clearly distinguishes mutations in RT-ddPCR. Droplet amplitude plots (n=1 per allele) illustrate how FAM fluorescence (X-axis) and VIC fluorescence (Y-axis) are diagnostic of templates matching the FAM probe (first column, blue droplets) or VIC probe (third column, green droplets) compared to templates with mutations in probe sequences (second column, orange droplets) and to droplets that lack template (fourth column, grey droplets) for each reaction (rows). Bar graphs (final column) show how consistent amplitudes are between specimens (average mean amplitude ± average standard deviation of 190-1900 positive droplets each, n=4 per allele). Note that in the 452/501 reaction, unlike the 417 and 484 reactions, the two probes have separate targets so droplets may show fluorescence from only one or the other target (droplets lower along the axes) or from both targets (droplets higher along the axes).
The researchers selected four SARS-CoV-2 lineages including the D614G, Beta, Gamma, and Kappa strains and investigated the amino acids located at RBD sites 417, 452, 484, 501. The genotyping results were compared to genome sequence for each sample to determine the sensitivity and specificity of the assay for each variant.
The authors tested a total of 2,657 SARS-CoV-2-positive specimens by TaqPath assay, 16 of which had a positive S-gene target failure (SGTF) result. RT-ddPCR was performed on these SGTF-samples and five non-SGTF samples.
Of the 16 SGTF-samples, one was identified as the Delta variant, whereas the remaining 15 samples were identified as the Omicron variant by a combination of droplet amplitudes that matched the published sequences for the SARS-CoV-2 Omicron variant. This finding was verified by whole genome sequencing (WGS), which also identified one Delta and 15 Omicron sequences.
![Template concentration does not affect assay accuracy. For each reaction (rows), composite droplet amplitude plots showing the highest-concentration sample of each allele (first column) and lowest-concentration dilution with >10 positive droplets of the same samples (second column) illustrate that amplitudes are diagnostic of template sequences despite wide differences in copy number per droplet. Circles or lines separating droplets from different alleles are for reference on the low-concentration plots. Bar graphs (final column) show this is consistent between specimens (mean amplitude ± standard deviation of highest [dark] and lowest [light] concentration of each specimen,](https://www.news-medical.net/images/news/ImageForNews_702086_16426567528165783.jpg)
Template concentration does not affect assay accuracy. For each reaction (rows), composite droplet amplitude plots showing the highest-concentration sample of each allele (first column) and lowest-concentration dilution with >10 positive droplets of the same samples (second column) illustrate that amplitudes are diagnostic of template sequences despite wide differences in copy number per droplet. Circles or lines separating droplets from different alleles are for reference on the low-concentration plots. Bar graphs (final column) show this is consistent between specimens (mean amplitude ± standard deviation of highest [dark] and lowest [light] concentration of each specimen, n=4 per allele).
Conclusions
The researchers designed an assay that was capable of successfully identifying SARS-CoV-2 variants, including the current dominant Omicron variant. The evolution of SARS-CoV-2 continues; therefore, new variants will continue to emerge over time. As a result, the identification, surveillance, and therapies against emerging SARS-CoV-2 variants should also evolve and advance in order to control the COVID-19 pandemic.
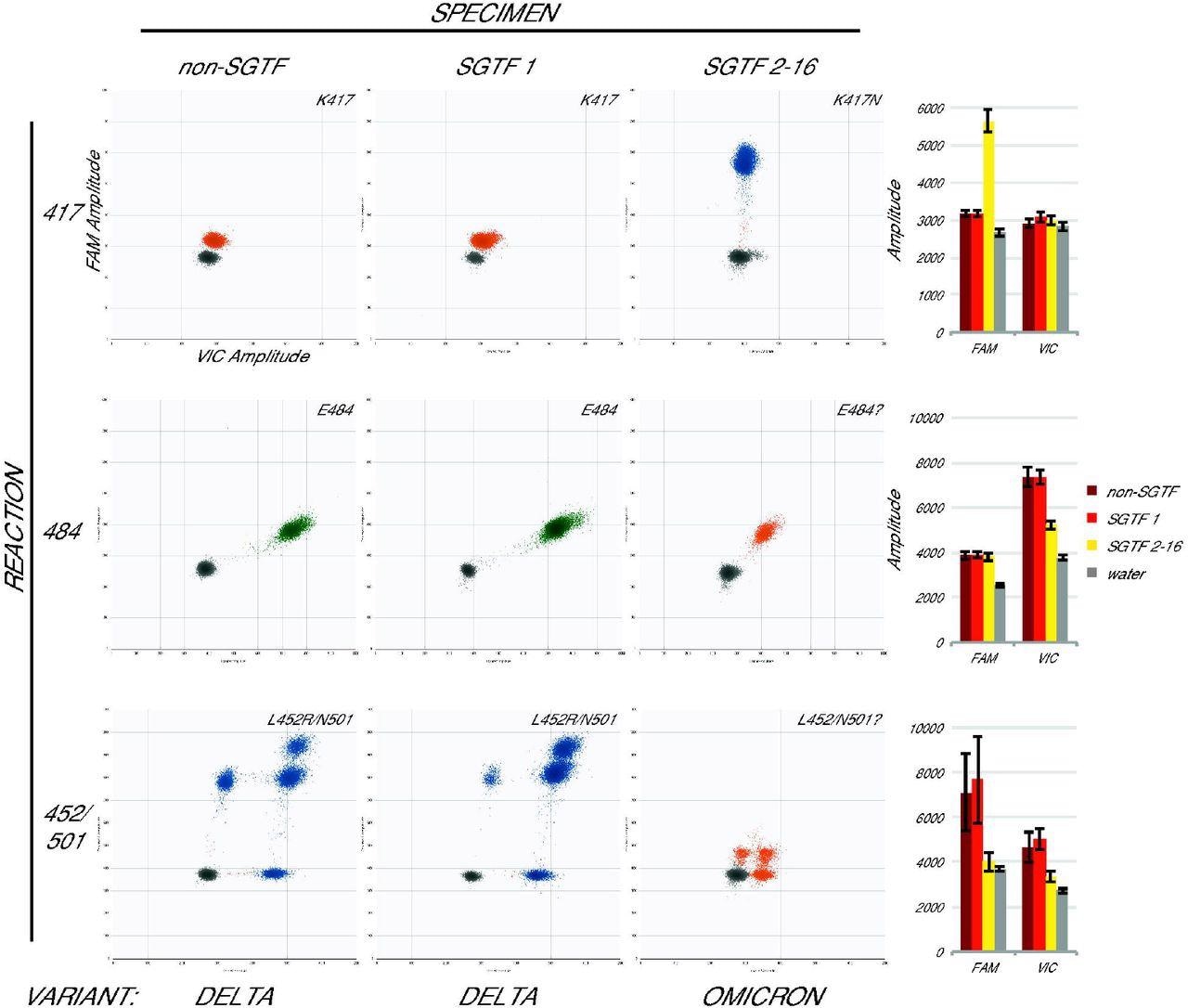
Omicron specimens are accurately identified with the assay. Droplet amplitude plots (n=1 per specimen type) illustrate the different assay results for three categories of newly-collected specimen: non-SGTF (first column), the first SGTF identified at UWVL (second column), and all subsequent SGTF (third column) for each reaction (rows). Bar graphs (final column) show average mean amplitude (± average standard deviation) for samples from each category (n=5, n=1, and n=7 respectively). Variant determination (bottom) based on the assay was confirmed in all cases by whole-genome sequencing.
The current technologies that are used for genomic sequencing of viral samples are advanced and provide accurate results of mutations. However, these experiments are often time-extensive and cannot be used to assist in clinical decision-making processes on whether mAb treatments would be effective for treating certain patients.
Taken together, the findings of the current study reveal that this new assay can easily and rapidly identify mutations in all previous and present SARS-CoV-2 variants and can be adapted to include future mutations.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Mills, M. G., Hajian, P., Bakhash, S. M., et al. (2022). Rapid and Accurate Identification of SARS-CoV-2 Omicron Variants Using Droplet Digital PCR (RT-ddPCR). medRxiv. doi:10.1101/2022.01.11.22268981. https://www.medrxiv.org/content/10.1101/2022.01.11.22268981v1.
- Peer reviewed and published scientific report.
Mills, Margaret G., Pooneh Hajian, Shah Mohamed Bakhash, Hong Xie, Derrek Mantzke, Haiying Zhu, Garrett A. Perchetti, et al. 2022. “Rapid and Accurate Identification of SARS-CoV-2 Omicron Variants Using Droplet Digital PCR (RT-DdPCR).” Journal of Clinical Virology 154 (September): 105218. https://doi.org/10.1016/j.jcv.2022.105218. https://www.sciencedirect.com/science/article/pii/S1386653222001512.