In an effort to better understand how the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) selectively targets and damages specific organs within the body upon infection, researchers have looked into how host cells interact with various SARS-CoV-2 proteins, including the spike, envelope, nucleocapsid, and membrane proteins.
 Study: Prediction and validation of host cleavage targets of SARS-CoV-2 3C-like protease. Image Credit: NIAID
Study: Prediction and validation of host cleavage targets of SARS-CoV-2 3C-like protease. Image Credit: NIAID

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Background
SARS-CoV-2 proteins can interact with various host biomolecules to initiate a wide range of damage to organs and tissues. More specifically, these host interactions have been found to cause suppression of the innate immune response and apoptosis, as well as the reprogramming of both transcription and translation processes within host cells.
Aside from the major structural proteins encoded by SARS-CoV-2, this virus also encodes for two proteases, including a papain-like protease (PLPro) and 3CLPro, the latter of which is often referred to as the main protease or Mpro. These two proteases are essential for viral replication and remain conserved across different types of coronaviruses; therefore, researchers have looked to these enzymes as novel targets for SARS-CoV-2 vaccines and therapeutics. In fact, recent Phase II/III clinical trials have found that the combination of PF-07323332, which is a 3CLPro inhibitor, with ritonavir reduced COVID-19-related hospitalization by up to 89%.
About the study
Despite the development of certain therapeutics to target SARS-CoV-2 proteases, their impact on the host proteome remains poorly understood. More specifically, there remains a lack of information available on the host cleavage targets of 3CLpro due to several limitations associated with current technologies.
In the current study, the researchers combine published cleavage efficiency data on the SARS-CoV 3CLPro, which is 96% similar to the 3CLpro of SARS-CoV-2, with genome-wide secondary structure analyses in order to identify and score predicted SARS-CoV and SARS-CoV-2 3CLPro cleavage sites across the human proteome. The algorithm used to identify and score these cleavage sites was based on data previously acquired by the researchers, wherein all FRET polypeptides between NSP4 and NSP5 were generated and modified with every possible single amino acid substitution.
With this information, the researchers assessed the cleavage efficiency of the SARS-CoV-2 3CLPro by comparing the observed fluorescence intensity between this protease and a reference cleavage sequence.
Taken together, this data allowed the researchers to develop an algorithm that generated a score for every possible cleavage site using a lookup table that multiplied the relative cleavage efficiency of every amino acid. This algorithm, which the researchers referred to as Sarsport1.0, then applied all multiplications to the entire human genome.
Both the precision and sensitivity of SARSport1.0 were also assessed in the current study. All scores, which were within the range of 1.31 to 0.04, were compared with published relative Kcat/Km values for each cleavage site.
Comparatively, the sensitivity of this algorithm to identify SARS-CoV-2 host protein targets was evaluated by calculating the scores for recently published and experimentally identified SARS-CoV-2 3CLPro cleavage targets. Out of the 117 previously identified targets, SARSport1.0 successfully identified 104 protein targets.
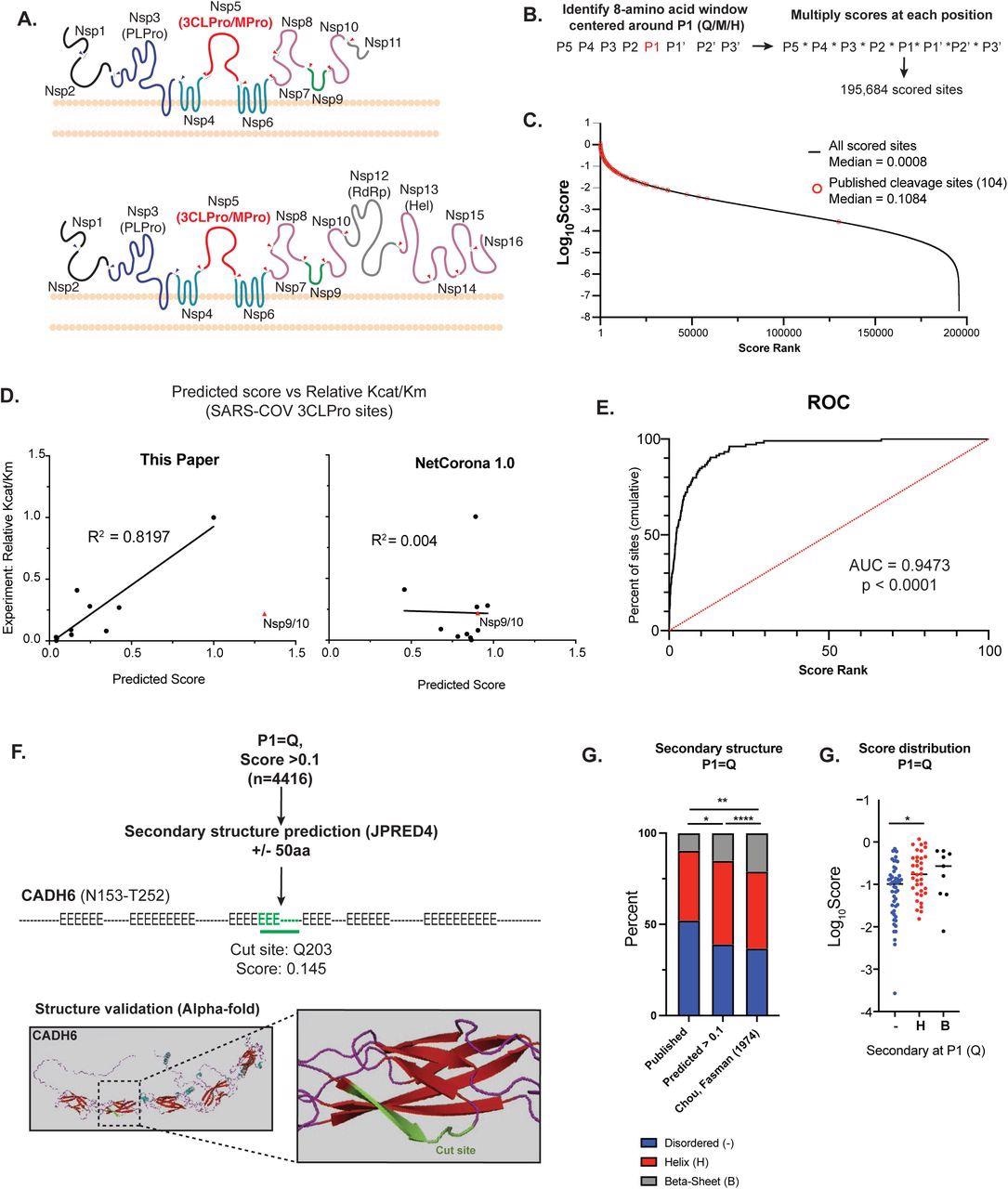
Bioinformatic prediction of SARS-COV2 3CL human protein targets A) Diagram of the endogenous function of the SARS-CoV-2 3C-like protease (3CLPro). 3CLPro cleaves at 11 sites within the 2 large polypeptides pp1a and pp1ab generated from the overlapping reading frames ORF1a and ORF1ab (respectively). Cleavage by 3CLPro, as well as the other viral protease papain-like protease (PLPro) liberates the nonstructural proteins (NSPs) that are required for viral transcription, replication, suppression of host immune responses and suppression of host gene expression. B) Work-flow for bioinformatic identification and scoring of putative 3CLPro cleavage sites within the human proteome. Scores for each position along the cleavage site (P5-P3’) were obtained from experimental data from SARS-CoV 3CLPro (Chuck, et al 2010). First P1 positions were identified (Q, M or H), and the overall cleavage score was generated by multiplying scores for each amino acid around P1. Within each 8-amino acid window, any position that contained an amino acid that scored a “ND” (no cleavage detected) in Chuck, et al resulted in a score of 0. Overall, 195,684 scored cleavage sites (>0) were detected across the human proteome. C) Distribution of all scores (Log10Score). Scores of published cleavage sites detected by our prediction are highlighted in red. D) Correlation of predicted score with experimentally-derived Kcat/Km values for SARS-COV 3CLPro (Grums-Tokars, 2008). Scores generated in this study are shown on the left, and scores generated by NetCorona1.0 (Kiemer, 2004). Shown on the right. For R2 calculations, the cleavage site between NSP9 and NSP10 for both graphs. E) Receiver operator curve analysis to assess the predictive power of bioinformatic scoring based on scores of published cleavage sites. Cumulative percentage of scores captured plotted vs score rank (highest score = 1 to lowest score = 100), and area under the curve (AUC) captured. 95% confidence interval determined by Wilson/Brown method. F) Secondary structure analysis of high scoring sites (>0.1) with P1 = Q. To increase secondary structure accuracy, a 100aa window centered around P1 (Q) was identified. Resulting 100aa peptides were analyses by JPRED4 to predict secondary structure ( “-“ = unstructured, “H” = alpha-helix, “E” = beta-sheet). When available, candidate cleavage sites were verified by alpha-Fold structure. Highlighted is a predicted site in Cadherin-6 (CADH6). G) Fraction of each P1 (Q) that lies in each type of secondary structure (unstructured, alpha-helix, beta-sheet). Comparisons are shown for predicted cleavage sites with score > 0.1 vs published cleavage sites (all scores) vs published secondary structure distribution of all glutamines (Q). Statistical analysis of secondary structure distribution calculated using Chi-squared goodness of fit. H) Score distribution of published cleavage sites (P1=Q), striated by secondary structure. Statistical analysis calculated by one-way ANOVA with Holm-Sidak multiple comparison test. For all statistics shown, * = p ≤ 0.05 , ** p ≤ 0.01, ***p ≤ 0.001, **** = p ≤ 0.00001
Assessing the in vitro cleavage potential of identified targets
The highly sensitive SARSport1.0 method developed in the current study was found to identify cleavage sites published in previous studies and identify several new predicted cleavage sites.
To validate the cleavage potential of these newly identified proteins, the researchers incubated purified 3CLPro with commercially available recombinant cadherin proteins, including CDH6 and CDH20, that had identical predicted sites.
This in vitro experiment confirmed that 3CLPro successfully and efficiently cleaved both CDH6 and CDH20 while also yielding predicted fragment sizes.
Incubation with NOTCH1 and 3CLPro also led to cleavage, resulting in the yielding of predicted fragments at their expected lengths. Additional protein targets of SVIL, UACA, and NOTCH2 were similarly validated in vitro.
Taken together, the findings from this experiment confirmed the ability of the SARS-CoV-2 3CLPro to cleave a wide range of host proteins. These cleavage events often lead to the generation of cytosolic protein fragments that ultimately get degraded by various endogenous pathways.
Cardiomyocyte-specific targets of 3CLPro
Sarcomeres often exhibit disorganization following SARS-CoV-2 infection; therefore, the researchers hypothesized that 3CLPro may have a role in degrading sarcomere proteins. To this end, SARSport1.0 was also applied to identify potential sarcomere targets of 3CLPro.
SARSport1.0 identified several potential sarcomere targets of 3CLPro, with the giant protein Obscurin (OBSCN) scoring as a highly likely target of this enzyme. In order to confirm the algorithm’s prediction, the researchers measured the levels of OBSCN in human induced pluripotent cell-derived cardiomyocytes (hiPSC-CMs) expressing 3CLpro. These experiments confirmed a marked reduction in OBSCN levels in these cells.
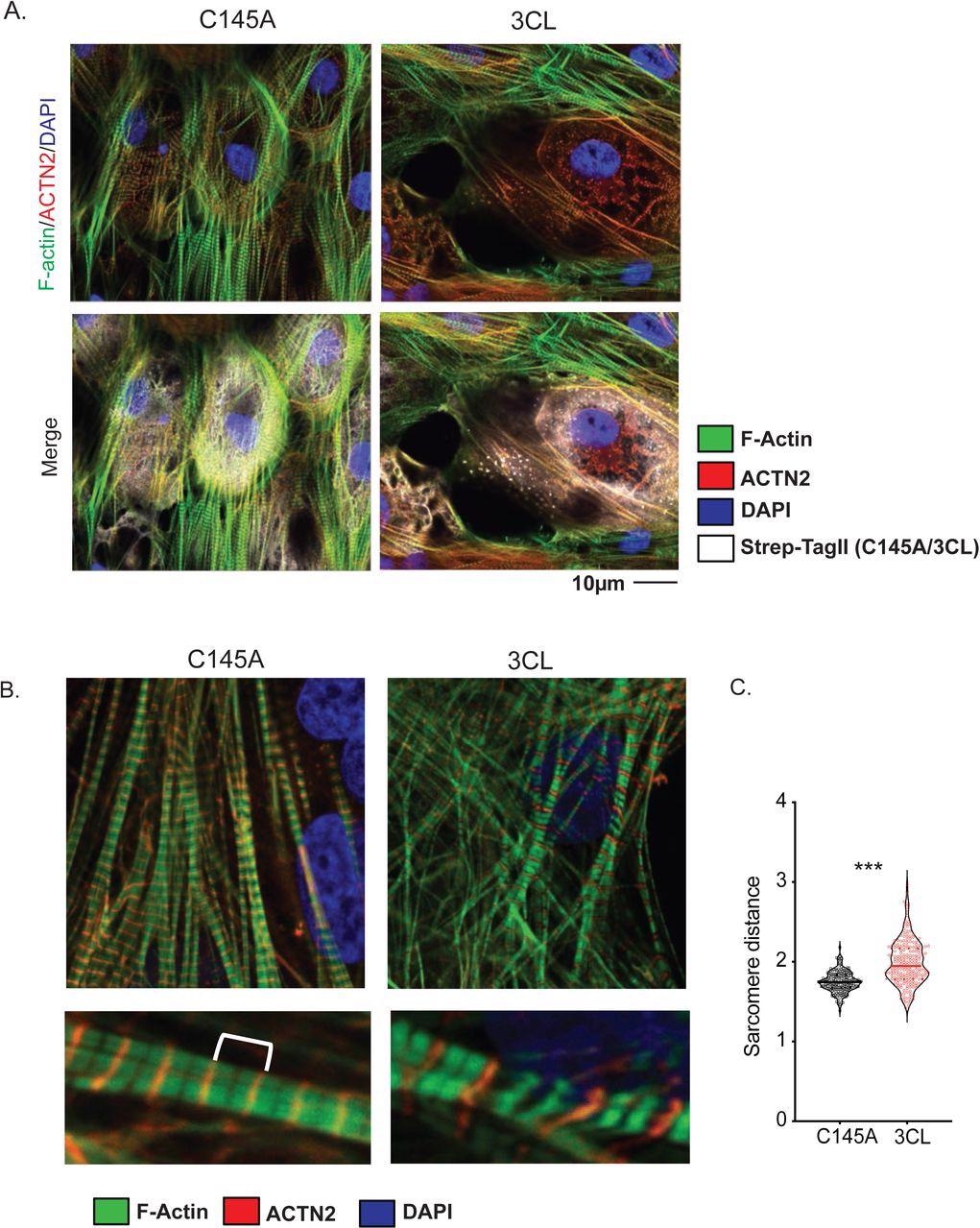
A) Sarcomere breakdown with overexpression of 3CLPro. hiPSC-CMs transduced with adenovirus overexpressing Strep-Tagged 3CLPro or catalytically inactive C145A control for 48h. Staining shows alpha-actinin (ACTN2), F-actin (phalloidin), and Strep-Tag. DAPI counterstain. B) Increased sarcomere length with overexpression of 3CLPro. Sarcomeres are stained with ACTN2 to mark Z-disks. An example image of sarcomeres with increased distance. C) Quantification of sarcomere distance. Sarcomere lengths (Z-disk to Z-disk length, as stained by ACTN2) were quantified for ∼200 sarcomeres (N=2/3 independent experiments? or only 1 exp with 200 sarcomeres?). Statistics shown by Student’s t-test (*** p = XXX). For all statistics shown, * = p ≤ 0.05 , ** p ≤ 0.01, ***p ≤ 0.001, **** = p ≤ 0.00001
Conclusions
The researchers of the current study provide several potential explanations for why the SARSport1.0 algorithm performed better than previous experimental approaches. For example, prior experimental approaches are limited to detecting proteins expressed in specific cells of interest.
Furthermore, certain proteomic approaches often rely on the ability to detect new protein fragments by mass spectrometry (MS). However, this analytical technique is often considered to be insensitive in its ability to detect protein fragments of a certain size and abundance. Furthermore, since MS requires the physical presence of cleaved products, the rapid degradation of many fragments can limit their detection by such approaches.
Previous studies have found that 3CLPro expression is one of the earliest events to occur during SARS-CoV-2 infection, which can lead to various effects on the host cell that subsequently alter the ensuing viral replication. The impact of early 3CLPro cleavage events could also contribute to the persistence of certain COVID-19 symptoms associated with long-COVID syndrome.
Taken together, the algorithm proposed in the current study overcomes many of the limitations associated with conventional in silico approaches to predicting cleavage events and products by 3CLPro. This highly sensitive and powerful tool could therefore be used to assist in virus-host interaction studies that often accompany early drug discovery processes.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Journal reference:
- Preliminary scientific report.
Prediction and validation of host cleavage targets of SARS-CoV-2 3C-like protease Nora Yucel, Silvia Marchiano, Evan Tchelepi, Germana Paterlini, Quentin McAfee, Nehaar Nimmagadda, Andy Ren, Sam Shi, Charles Murry, Zoltan Arany bioRxiv 2022.01.17.476677; doi: https://doi.org/10.1101/2022.01.17.476677, https://www.biorxiv.org/content/10.1101/2022.01.17.476677v1