As of January 23, 2022, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected over 350 million people worldwide and claimed the lives of over 5.59 million. A new study posted to the medRxiv* preprint server describes the emergence of new SARS-CoV-2 variants in three immunocompromised patients.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
With the rollout of several effective vaccines against the coronavirus disease 2019 (COVID-19), public health officials anticipated that rapid vaccination coverage would lead to population immunity and the end of the current pandemic. However, delays and shortfalls in the supply of vaccines to many regions, along with vaccine skepticism, have led to the emergence of several SARS-CoV-2 variants that are associated with higher transmissibility and immune escape capabilities. These new strains of SARS-CoV-2 have driven successive resurgences of the virus that have caused new infections, reinfections, and breakthrough infections.
Many previous studies have described chronic SARS-CoV-2 infections in immunocompromised patients due to their inability to clear the virus completely. The prolonged presence of SARS-CoV-2 in the body allows the viral genome to change and adapt to environmental threats, including antibodies used to treat such patients.
This is a fertile source for novel SARS-CoV-2 variants, especially those with antibody escape capability from antibodies elicited by both vaccines and natural infection. Mutations in the spike gene can lead to changes in the spike protein that modify the antibody epitopes and subsequently prevent or impair spike binding by anti-spike antibodies. As a result, neutralizing antibodies fail to act against these novel variants.
Many of these SARS-CoV-2 variants, including the Alpha, Beta, Delta, and Omicron variants, have multiple spike mutations that are associated with antibody escape from neutralizing antibodies that are active against the wild-type virus. Neutralizing activity is a correlate of protective immunity against the virus, as well as viral clearance and non-fatal disease; therefore, any reduction in this activity is significant.
The current study describes the emergence of novel mutations in three patients with chronic SARS-CoV-2 infection because of advanced human immunodeficiency virus (HIV) infection.
Study findings
The first patient was admitted to the hospital with breathlessness along with weight loss, fevers, and night sweats. A polymerase chain reaction (PCR) test of the combined nose and throat swabs (CNTS) was positive for SARS-CoV-2 and the patient was also diagnosed to have HIV-1. Military tuberculosis was also present, and the patient was treated for all these conditions. Two months later, a repeat CNTS was negative for SARS-CoV-2 ribonucleic acid (RNA).
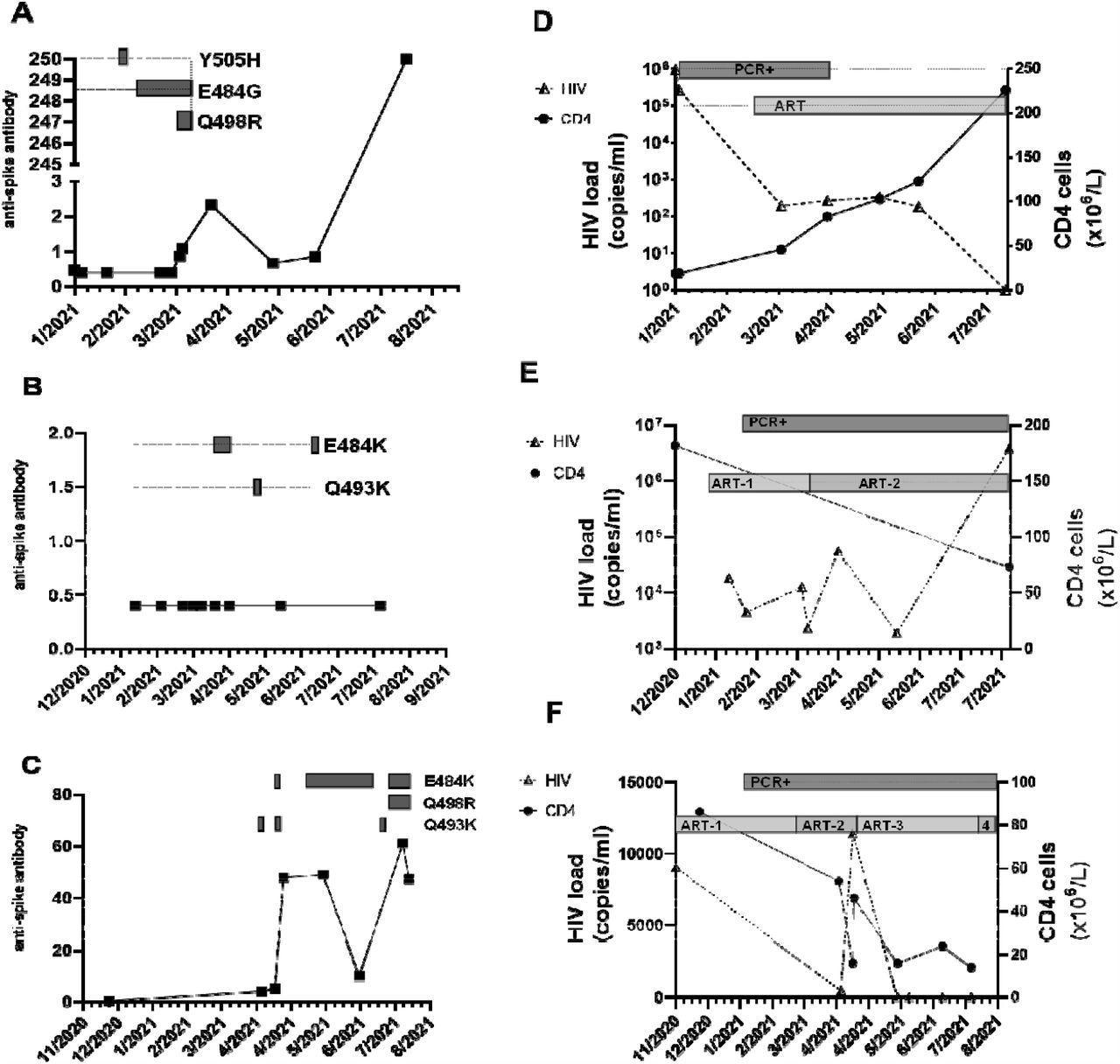
Serial CNTS samples from (a; d) patient A; (b; e) patient Band (c; f) patient C. Anti-spike IgG was measured using commercial assay (Roche Elecys anti-SARS-COV-2 anti –Spike antibody), units of measure are arbitrary, cut-off for positive result indicated by dashed horizontal line. Amino acid changes in the receptor binding domain (RBD) of the spike protein at locations previously associated with variants of concern (VOC) that arose over the same time period are displayed as boxes along the dotted horizontal line. Amino acid changes are either seen at the consensus level or are part of a mixed viral population. In addition, the alpha lineage mutation N501Y was present in all samples. HIV-1 viral load was measured using the Roche Cobas 8800, and CD4 cell count were measured and the SARS-COV-2 RT-PCR positivity and ART regimen over the same time is also displayed. (d) ART: Trimeq (DTG/ABC/3TC)+DTG; (e) ART-1: Bictarvy (BTG/TAF/ETC), ART-2: TAF/ETC/DRV/c; (f) ART-1: TDF/ETC/DRV/r; ART -2 : TAF/ETC/DRV/c; ART-3: TAF/ETC/DTG; ART-4: TDF/ETC/DTG
The second patient, who had previously been diagnosed with diffuse large B-cell lymphoma and HIV-1, was admitted as a result of a urinary tract infection. The patient was poorly compliant with antiretroviral therapy (ART) for HIV, which had been diagnosed two decades before. At four weeks from admission, a CNTS was positive for SARS-CoV-2.
This second patient was put on oxygen due to breathlessness and remained positive for SARS-CoV-2 thereafter.
The third patient had a ten-year history of HIV-1 with intermittent ART. Presenting with cough, fever, and myalgia, the diagnosis of COVID-19 was made upon admission. While the symptoms subsided soon afterward, every weekly CNTS has been positive for SARS-CoV-2.
All patient samples yielded large amounts of viral RNA from the CNTS, which corresponded to a cycle threshold (Ct) of 30 that is considered to be a moderate-to-high viral load. In all cases, the infecting variant was the Alpha SARS-CoV-2, which consists of an N501Y mutation that improves the binding affinity of the viral receptor-binding domain (RBD) for the host cell angiotensin-converting enzyme 2 (ACE2) receptor by ten-fold.
When the viral RNA from these patients was sequenced by next-generation sequencing (NGS), numerous mutations in the SARS-CoV-2 spike RBD were detected. Some of these mutations persisted over time, probably because they were selected for by the presence of antibodies to earlier strains.
All patient samples were also found to harbor a mutation at E484 within the receptor-binding motif (RBM) of the RBD, a position notorious for immune escape. Some of these mutations, such as E484K and E484G, also increase the binding affinity of the RBD for ACE2.
The anti-spike antibodies in the serum of the third patient were at detectable levels; however, no activity was seen with the wild-type virus. Successful virus isolation was achieved from the third patient.
Implications
The current study demonstrates the evolution of SARS-CoV-2 within immunocompromised hosts harboring chronic infection and its adaptation through novel mutations that emerged over time. The persistence of some, though not all, of the mutations, indicates that SARS-CoV-2 continued to replicate actively within the patients as a result of the immune deficit that led to a failure to clear the virus from the body.
“These findings provide further evidence that in the setting of immunosuppression, individuals can have chronic SARS-CoV-2 infection providing an opportunity for the generation of novel variants.”
These mutations appear to promote higher transmissibility and could also facilitate antibody escape. The combined presence of N501Y and E484K in the SARS-CoV-2 Beta variant was associated with the highest reduction in neutralizing capacity by currently available natural, vaccine-induced, and therapeutic monoclonal antibodies seen so far.
The presence of the Q498R mutation, in combination with N501Y, is expected to markedly promote binding affinity for the ACE2 receptor. When all three mutations occur, as was seen in the first and third patients, their cumulative effects are expected to enhance RBD-ACE2 binding 50-fold as compared to the wild-type virus.
Such large increases in transmissibility are worrying and could indicate that the variant is also capable of efficiently evading antibody-mediated neutralization. Further studies are therefore needed to characterize the impact of these mutations.
However, it is already known that other mutations, such as Q493K in the RBD that is also found in the Omicron variant, along with Q498R and Y505H, are associated with resistance to neutralizing antibodies and have been previously identified in immunocompromised hosts. The possible causes of these mutations could include the very low CD4 cell counts, absence of a neutralizing antibody response, or the increased receptor binding that allowed certain mutations to be positively selected.
“CD4 T-cells are central to both neutralizing antibody and cytotoxic T-cell activity in acute infection.”
The lack of adequate CD4 cell counts could be responsible for the poor neutralizing antibody response since the latter depends on activation by T-cells and because CD4 T-cells are linked with recovery from infection with this virus and faster viral clearance. However, hen the CD4 cell response is absent or significantly delayed, a fatal or severe outcome is more likely. In contrast, the patients in this study had chronic yet asymptomatic SARS-CoV-2 infection, perhaps due to the contribution of other components of adaptive immunity.
The lack of neutralization in the third patient, even with detectable anti-spike antibody titers, shows that the functional effects of antibodies, rather than their magnitude alone, must be measured, especially in immunocompromised patients. Much more research is required to discern the relationships between the deficiency of different immune components and viral clearance in this and other infections.
Meanwhile, it is obvious that patients with chronic SARS-CoV-2 infection must be monitored closely to prevent the emergence of new mutations that can facilitate escape from vaccine-induced antibodies. In this case, genome sequencing data was used to identify the presence of replicating virus particles, thus triggering proper containment measures. This would indicate the need for such sequencing tools to be made available in diagnostic centers in order to prevent and contain such outbreaks.
A significant proportion of the sub-Saharan African population remains unvaccinated. The high HIV caseload, about a third of whom are not on ART and thus chronically immunosuppressed, highlights the threat that these individuals could promote further mutation of SARS-CoV-2. Taken together, these factors support the need to make vaccination a global program, rather than regional.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Riddell, A. C., Kele, B., Harris, K., et al. (2022). Generation of Novel SARS-Cov-2 Variants on B.1.1.7 Lineage in Three Patients with Advanced HIV Disease. medRxiv. doi:10.1101/2022.01.14.21267836. https://www.medrxiv.org/content/10.1101/2022.01.14.21267836v1.
- Peer reviewed and published scientific report.
Riddell, Anna C, Beatrix Kele, Kathryn Harris, Jon Bible, Maurice Murphy, Subathira Dakshina, Nathaniel Storey, et al. 2022. “Generation of Novel Severe Acute Respiratory Syndrome Coronavirus 2 Variants on the B.1.1.7 Lineage in 3 Patients with Advanced Human Immunodeficiency Virus-1 Disease.” Clinical Infectious Diseases 75 (11): 2016–18. https://doi.org/10.1093/cid/ciac409. https://academic.oup.com/cid/article/75/11/2016/6591841.