
 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
Although the SARS-CoV-2 Omicron variant has higher infectivity and immune evasion induced by previous infection or vaccination, the severity of COVID-19 in convalescent and vaccinated individuals is lower, indicating the protection by previously primed SARS-CoV-2-specific T cells cross-recognition of Omicron. However, comparative in-depths studies regarding the SARS-CoV-2-specific T cell epitope repertoire targeted by vaccination compared to infection-induced T cell responses are lacking.
About the study
In the present study, the researchers analyzed SARS-CoV-2-specific T cell responses in 19 individuals who recovered from SARS-CoV-2 infection, 16 participants who received two doses of Pfizer/BioNTech mRNA vaccine, and seven individuals who were mRNA booster vaccinated.
The team initially mapped the overall SARS-CoV-2-specific CD8+ T cell response. Further, they evaluated a set of 43 previously described immunodominant SARS-CoV-2-specific CD8+ T cell epitopes restricted by common human leukocyte antigen (HLA) class I alleles in epitope-specific T cell cultures after cytokine staining.
The CD8+ T cell responses in convalescents versus vaccinees were analyzed using overlapping peptides spanning the whole spike protein and an in-silico analysis. Further, the CD4+ T cell responses and the effect of boosting convalescents and two-dose vaccinated individuals were determined using overlapping spike peptides spanning the whole spike protein.
Finally, the researchers analyzed T cell response targeting highly conserved selective sweep regions in SARS-CoV-2 among convalescents compared to vaccinated individuals.
Results
The results demonstrated that convalescent individuals had CD8+ T cell responses against most of the epitopes of all SARS-CoV-2 proteins; however, spike (S)-specific responses were not dominant. In contrast, the SARS-CoV-2 vaccinees had CD8+ T cells responses targeted against the S epitopes. Some of the CD8+ T cells responses were also directed towards non-S epitopes such as HLA-B*07/N, for which T cells have previously shown cross-recognition against common cold coronaviruses. In the SARS-CoV-2 vaccinees, individual S-specific CD8+ T cells epitopes were targeted the most and had a broad range of S-specific CD8+ T cell repertoire compared to convalescent individuals.
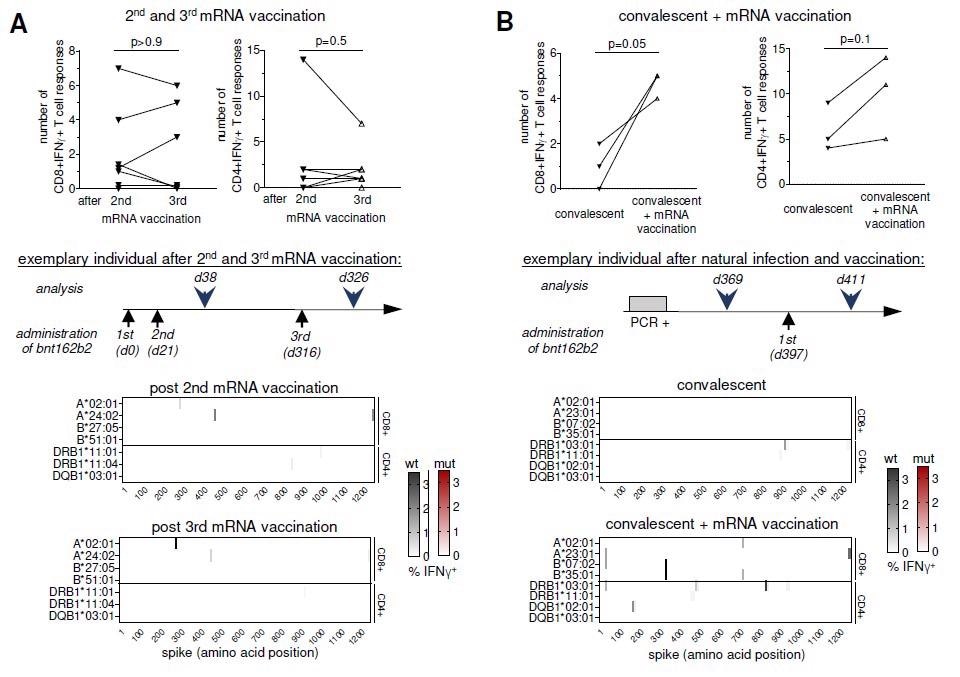
Boosted vaccine- and infection-induced spike-specific CD8+ and CD4+ T cell responses. Number, location and percentages of spike-specific CD8+ and CD4+ T cell responses to overlapping peptides (OLP) that are detectable in SARS-CoV-2 vaccinees after the 2nd versus after the 3rd dose (A) of Pfizer/BioNTech mRNA vaccine (measured 2-4 weeks after vaccination) and in SARS-CoV-2 convalescents who subsequently received a single dose (B) of Pfizer/BioNTech mRNA boost vaccination (measured 2 weeks after vaccination) are depicted. Targeted epitopes with sequence variations in omicron are marked in red. statistical analysis was performed with paired t test.
The comparison between corresponding viral sequences of the SARS-CoV-2 ancestral and Omicron variants showed that viral variation affected only a single CD8+ T cell epitope.
SARS-CoV-2-vaccinated individuals had significantly broader S-specific CD8+ T cell responses repertoire than the convalescents. While several HLA class I alleles restricted five or more S-specific CD8+ T cell epitopes in SARS-CoV-2 vaccinees, no HLA class I allele restricted more than two S-specific CD8+ T cell epitopes in convalescents. In addition, vaccinated individuals showed a higher number of S-specific CD8+ T cell responses per individual compared to those who had SARS-CoV-2 previously.
In contrast to CD8+ T cell responses, vaccinees showed a more limited repertoire of targeted S-specific CD4+ T cell response epitopes than the SARS-CoV-2 convalescent individuals. Further, fewer S-specific CD4+ T cell epitopes were restricted by single HLA class II alleles, and fewer CD4+ T cell responses were detectable per individual in SARS-CoV-2-vaccinated individuals compared to convalescent individuals. Individual and population-based CD4+ T cell epitopes showed limited S-specific CD4+ T cell repertoire in SARS-CoV-2 vaccinees. High conservation between SARS-CoV-2 ancestral and Omicron variants was observed among the fewer targeted S-specific CD4+ T cell epitopes in SARS-CoV-2 vaccinees, as well as convalescent individuals.
Following mRNA booster vaccination, the participants had a similarly broad and cross-reactive S-specific CD8+ T cell repertoire and similarly limited; however, cross-reactive S-specific CD4+ T cell repertoire to that of SARS-CoV-2 convalescents and the two-dose mRNA vaccinated individuals. Convalescents had a broader S-specific CD8+ T cell repertoire after mRNA booster vaccination, whereas CD4+ T cell repertoire was similar before and after booster vaccination. The identified CD8+ and CD4+ T cell responses targeted epitopes were conserved in Omicron.
Highly conserved selective sweep regions in SARS-CoV-2 mediate per definition an evolutionary advantage in T cell responses in convalescents and vaccinees. The four different selective sweep regions that have been described to date exhibited a high degree of amino acid homology among the already evolved SARS-CoV-2 variants of concern (VOCs). Furthermore, vaccinated individuals demonstrated S-specific CD8+ T cell responses targeting epitopes within the highly conserved selective sweep regions compared to SARS-CoV-2 convalescents. However, a similar highly conserved selective sweep region targeted S-specific CD4+ T cell response was not evident after vaccination.
Conclusions
The study findings indicated that while convalescents target a wide range of SARS-CoV-2-specific CD8+ T cell epitopes over the complete SARS-CoV-2 proteome, S-specific CD8+ T cell responses were not dominant. Opposite to the CD4+ T cell response, vaccinees had CD8+ T cell responses targeted to a broader repertoire of highly conserved S-specific CD8+ T cell epitopes resulting in high cross-recognition potential. The mRNA booster vaccination in convalescents broadened S-specific CD8+ T cell response. Further, the CD8+ and CD4+ T cell responses in the SARS-CoV-2 convalescent and vaccinated individuals target epitopes that are highly conserved within ancestral SARS-CoV-2, Omicron, and possibly in future emerging SARS-CoV-2 VOCs.
Overall, the study findings underscore the significance of mRNA booster vaccine-induced S-specific CD8+ T cell responses in mitigating emerging SARS-CoV-2 VOCs, including Omicron, as well as the importance of mRNA booster vaccination in SARS-CoV-2 convalescent individuals.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
SARS-CoV-2 specific T cells induced by both SARS-CoV-2 infection and mRNA vaccination broadly cross-recognize omicron, Julia Lang-Meli, Hendrik Luxenburger, Katharina Wild, Vivien Karl, Valerie Oberhardt, Elahe Salimi Alizei, Anne Graeser, Matthias Reinscheid, Natascha Roehlen, Sebastian Giese, Kevin Ciminski, Veronika Götz, Dietrich August, Siegbert Rieg, Cornelius F. Waller, Tobias Wengenmayer, Dawid Staudacher, Bertram Bengsch, Georg Kochs, Martin Schwemmle, Florian Emmerich, Tobias Boettler, Robert Thimme, Maike Hofmann, Christoph Neumann-Haefelin, Research Square, 2022, doi: https://doi.org/10.21203/rs.3.rs-1269004/v1, https://www.researchsquare.com/article/rs-1269004/v1
- Peer reviewed and published scientific report.
Lang-Meli, Julia, Hendrik Luxenburger, Katharina Wild, Vivien Karl, Valerie Oberhardt, Elahe Salimi Alizei, Anne Graeser, et al. 2022. “SARS-CoV-2-Specific T-Cell Epitope Repertoire in Convalescent and MRNA-Vaccinated Individuals.” Nature Microbiology 7 (5): 675–79. https://doi.org/10.1038/s41564-022-01106-y. https://www.nature.com/articles/s41564-022-01106-y.