The ongoing coronavirus disease 2019 (COVID-19) pandemic has been caused by a novel coronavirus, namely, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). This highly contagious RNA virus belongs to the family Coronaviridae of the genus Betacoronavirus. The SARS-CoV-2 infection causes a wide array of clinical symptoms, including mild to severe pneumonia-like symptoms. In addition, many individuals are also asymptomatically infected by the virus.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
Several studies have reported that anti-SARS-CoV-2 antibodies and T cell responses are correlated with milder disease and accelerated viral clearance. These studies have also revealed that SARS-CoV-2–specific CD4+ and CD8+ T cell responses control COVID-19 infection. Researchers have adopted varied approaches to evaluate virus-specific T cell responses in acute, convalescent, and severe patients to understand antigen specificity.
Prior studies have used HLA to predict peptide megapools (MP) of the SARS-CoV-2 proteome. These studies have highlighted the presence of SARS-CoV-2-specific CD4+ T cell responses in all COVID-19 patients. The majority of the CD4+ T cell reactivity found in these studies was directed towards the SARS-CoV-2 spike (S), membrane (M), and nucleocapsid (N) proteins. In addition, researchers have observed that CD4+ T cell responses to the SARS-CoV-2 S or N proteins were correlated with the amount of the anti-SARS-CoV-2 neutralizing antibodies present in COVID-19 convalescent patients. However, the SARS-CoV-2 M protein has been associated with the evasion of protective immune responses by controlling innate immune responses. In this context, the M protein is also linked with hampering interferon (IFN) signaling pathways and disturbing the production of type I and III IFN. Therefore, it is imperative to understand the molecular mechanism behind the SARS-CoV-2-specific CD4+ T cell response against the SARS CoV-2 structural proteins. This understanding would help the development of effective COVID-19 therapeutics and vaccines.
A New Study
A new study, published on the medRxiv* preprint server, focused on the production of human CD4+ T cell lines (TCLs), reactive to either S or M protein, and analyzed if the S and M structural proteins had intrinsic differences in driving antigen-specific CD4+ T cell responses. In this study, researchers used T cells from six unexposed naïve healthy volunteers prior to the COVID-19 pandemic.
Scientists observed that both the S and M proteins of SARS-CoV-2 drove a strikingly distinct molecular signature in TCLs under neutral conditions. They reported that S-specific responses were not markedly distinguishable from responses induced by other viruses, e.g., cytomegalovirus. However, the M protein-specific CD4+ TCLs had a transcriptomic signature, exhibited by a distinct suppression of STAT1-IFRs-interferon pathway signaling.
Previous studies revealed that CD4+ T cells are strongly associated with reducing COVID-19 disease severity compared to B cells and CD8+ T cells. Unsurprisingly, the absence of SARS-CoV-2-specific CD4+ T cells has been linked with severe or fatal COVID-19. Several studies have shown that patients infected with mild symptoms and those who recovered without treatment revealed a high level of SARS-CoV-2-specific CD4+ T cell responses to S and N proteins.
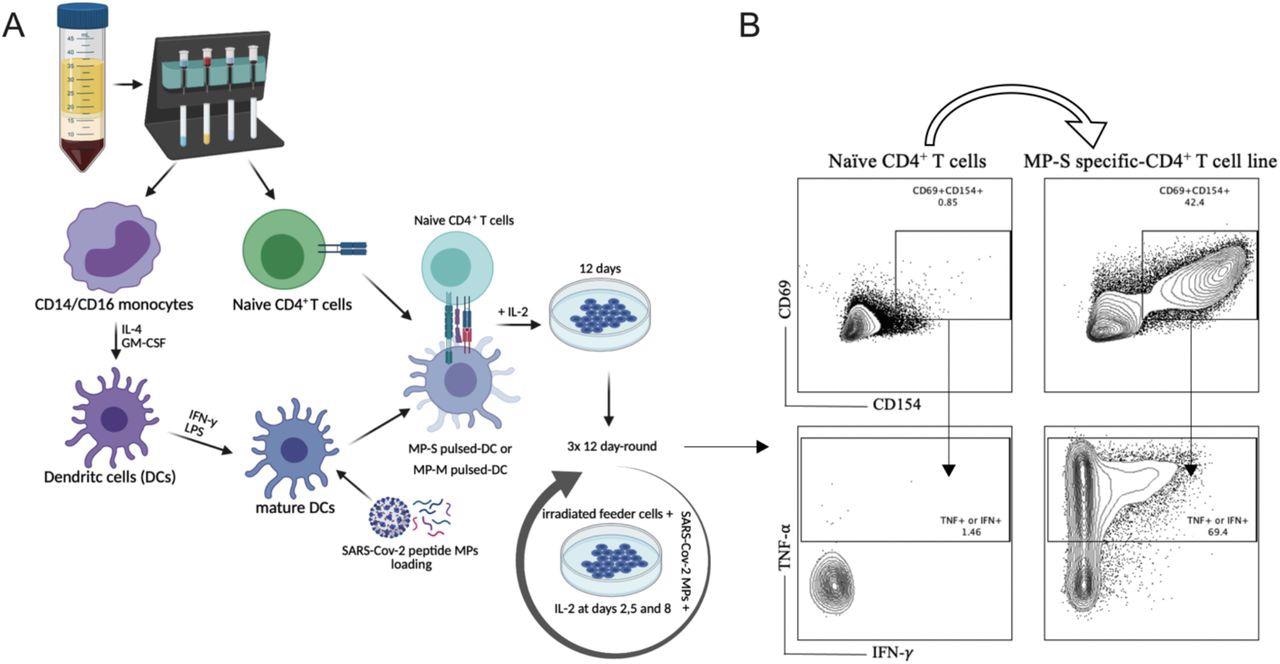
SARS-Cov-2 antigen-specific CD4+ T-cell line generation from healthy donor PBMC. Panel A demonstrates the methodology used for the generation of the various TCLs beginning with healthy donor naive T-cells and driven by SARS-Cov-2 structural spike (S) and membrane (M) protein-based peptide megapools. Panel B shows a representative flow cytometric analysis of MP-S-specific CD4+ TCLs expressing CD69+CD154+ in comparison with their respective naïve CD4+ T cells. Moreover, upon stimulation with their respective MPs, the vast majority of CD69+CD154+ MP-S specific-CD4+ T cell lines were either IFN-y+ or TNF-a+.
The current study used single-cell transcriptomes of human CD4+ TCLs reactive to either S protein or M protein and reported that SARS-CoV-2 M protein-reactive CD4+ TCLs, in comparison to the S protein, activated higher levels of the inflammatory genes, such as FOS, JUNB, and lower levels of IFN-stimulated genes (ISGs) (e.g., ISG15, IFI6, IFI35), and interferon regulatory factors (e.g., IRF7). Scientists explained that viral recognition induces IFN production, which subsequently activates the transcription of ISGs. Activation of ISGs engages various antiviral functions. One of the primary roles of ISG is the regulation of type 1 interferons.
Among the different ISGs, ubiquitin-like protein ISG15 is most robustly and rapidly induced, associated with inhibition of viral replication and modulating host immunity. In this study, the molecular signature of SARS-CoV-2 M protein-reactive CD4+ TCLs was characterized by suppression in the interferon pathways. This suppression has been genetically connected with the transcriptional profile of severe COVID-19 infection. The findings of this study are in line with previous reports, which revealed that CD4+ T cells from patients with severe COVID-19 showed reduced expression of interferon-induced genes, including IFIT1, IFIT2, IFIT3, and IFITM1.
Conclusion
The authors stated that the mechanisms behind the peptide megapools of M and S proteins associated with different responses of naïve CD4+ T cells should be studied in the future. Additionally, more research is required to understand if M protein-driven dysregulation of interferon signaling pathway in the adaptive immunity is similar to the mechanisms associated with pattern recognition receptors (PRRs) downstream molecules of innate cells. The current study strongly suggested a potential connection between the antigen specificity of the reactive CD4+ T cells to SARS-CoV-2 and the development of a functional adaptive immune response. This study also indicated that the SARS-CoV-2 M protein could also be used as an antigenic target to produce COVID-19 therapeutics.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Gazzinelli-Guimaraes, H. P. et al. (2022) Antigenic determinants of SARS-CoV-2-specific CD4+ T cell lines reveals M protein-driven dysregulation of interferon signalling. medRxiv, 2022.01.20.22269491; doi: https://doi.org/10.1101/2022.01.20.22269491, https://www.medrxiv.org/content/10.1101/2022.01.20.22269491v1
- Peer reviewed and published scientific report.
Gazzinelli-Guimaraes, Pedro H., Gayatri Sanku, Alessandro Sette, Daniela Weiskopf, Paul Schaughency, Justin Lack, and Thomas B. Nutman. 2022. “Antigenic Determinants of SARS-CoV-2-Specific CD4+ T Cell Lines Reveals M Protein-Driven Dysregulation of Interferon Signaling.” Frontiers in Immunology 13 (June). https://doi.org/10.3389/fimmu.2022.883159. https://www.frontiersin.org/articles/10.3389/fimmu.2022.883159/full.