
 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
MERS-CoV belongs to the linage C of beta-coronaviruses (CoV) (merbecoviruses) and is associated with a higher mortality rate. MERS-CoV has been found in various animal species including bats, camels, and hedgehogs.
Dipeptidyl peptidase 4 (DPP4) is the functional receptor of MERS-CoV and several other CoVs in bat. Sufficient information regarding the closest relatives of MERS-CoV, including NeoCoV, PDF-2180-CoV, HKU5-CoV, and hedgehog CoV EriCoV-HKU31, is not yet available. The lack of information about bat CoVs target receptors has hindered the understanding of these high-consequence pathogens.
About the study
In the present study, researchers explore the receptors of MERS-CoV-like viruses in bats. The relationship between merbecoviruses was determined through phylogenetic analysis of a list of animal and human coronaviruses sequences.
The divergence of the spike (S) S1 encoding region in NeoCoV and PDF-2180-CoV compared to HKU4-CoV was assessed by a sequence similarity plot analysis using MERS-CoV as a query. Further, the affinity of marbecoviruses towards human DPP4 (hDPP4) was determined using a pseudovirus entry assay.
The affinity of NeoCoV and PDF-2180-CoV viruses towards bat ACE2 was evaluated by screening a bat ACE2 cell library. The two viruses' affinity towards the bat ACE2 and human ACE2 (hACE2) was assessed by a dual split protein (DSP)-based fusion assay. The species-specific utilization of ACE2 by NeoCoV and PDF-2180-CoV was determined by a live-cell binding assay using cells expressing various bat ACE2.
The binding affinity of both PDF-2180-CoV and NeoCoV to ACE2 was determined using Bio-Layer Interferometry (BLI) analysis and enzyme-linked immunosorbent assay (ELISA). The binding affinity was also verified by competitive neutralization assays using soluble viral S1-CTD-hFc or ACE2-ectodomain proteins.
The molecular basis of the virus-ACE2 binding was determined by conducting three-dimensional (3D) structural investigations of the PDF-2180-CoV and NeoCoV receptor-binding domain (RBD)-Bat37ACE2 complex.
Study findings
Both NeoCoV and PDF-2180-CoV were found to form a sister clade with MERS-CoV in the phylogenetic analysis. However, the NeoCoV and PDF-2180-CoV amino acid sequence of the SS1 subunit was distinct from MERS-CoV and closely resembled the hedgehog coronaviruses (EriCoVs) in the analysis.
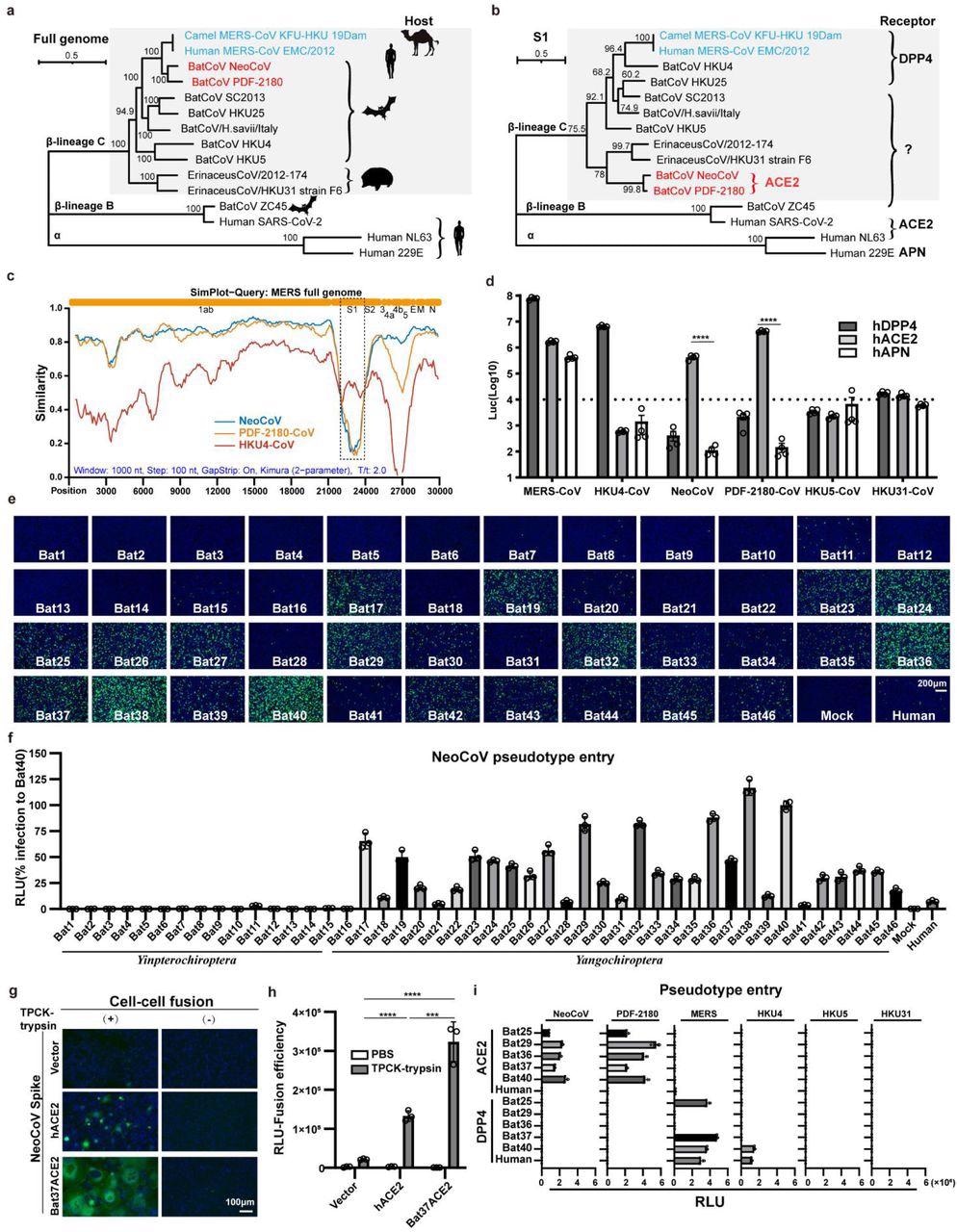
A clade of bat merbecoviruses can use ACE2 but not DPP4 for efficient entry. a-b, Phylogenetical analysis of merbecoviruses (gray) based on whole genomic sequences (a) and S1 amino acid sequences (b). NL63 and 229E were set as outgroups. Hosts and receptor usage were indicated. c, Simplot analysis showing the whole genome similarity of three merbecoviruses compared with MERS-CoV. The regions that encode MERS-CoV proteins were indicated on the top. Dashed box: S1 divergent region. d, Entry efficiency of six merbecoviruses in 293T cells stably expressing hACE2, hDPP4, or hAPN. e-f, Entry efficiency of NeoCoV in cells expressing ACE2 from different bats. EGFP intensity (e); firefly luciferase activity (f). g-h, Cell-cell fusion assay based on dual-split proteins showing the NeoCoV spike protein mediated fusion in BHK-21 cells expressing indicated receptors. EGFP intensity (g), live-cell Renilla luciferase activity (h). i, Entry efficiency of six merbecoviruses in 293T cells stably expressing the indicated bat ACE2 or DPP4. Mean±SEM for d, i; Mean±SD for f, and h.(n=3). RLU: relative light unit.
HKU4-CoV and MERS-CoV caused a significant infection of 293T-hDPP4 in the pseudovirus entry assay. Unexpectedly, a substantial increase of NeoCoV and PDF-2180-CoV entry of 293T-hACE2 was noted in the pseudovirus assay.
Similarly, an increase in the entry of NeoCoV and PDF-2180-CoV in bat ACE2 cells was observed during the bat ACE2 cell library screening.
The closely related NeoCoV and PDF-2180-CoV significantly bind with certain bat ACE2, particularly those belonging to the Yangochiroptera group and, to a lesser extent, in hACE2 for cell entry according to the DSP-based fusion assay. Further, the PDF-2180-CoV and NeoCoV's S1 subunit carboxyl-terminal domains (S1-CTD) enable the species-specific and high-affinity ACE2 binding.
The cryo-electron microscopy analysis of NeoCoV RBD bound with the Pipistrellus pipistrellus ACE2 protein demonstrated a new protein-glycan interaction by the glycosylation at N54. This analysis also demonstrated that NeoCoV binds to the apical side surface of ACE2, which differs from other known hACE2-binding viruses such as NL63 or the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
A molecular determinant around the viral binding interface, particularly close to the residue Asp338, limits the NeoCoV-hACE2 binding. However, following a T510F mutation on the receptor-binding motif (RBM), NeoCoV substantially infects ACE2 expressing human cells. Moreover, neutralizing antibodies against SARS-CoV-2 or MERS-CoV elicited by the current coronavirus disease 2019 (COVID-19) vaccines did not exhibit cross-neutralization with the NeoCoV infection.
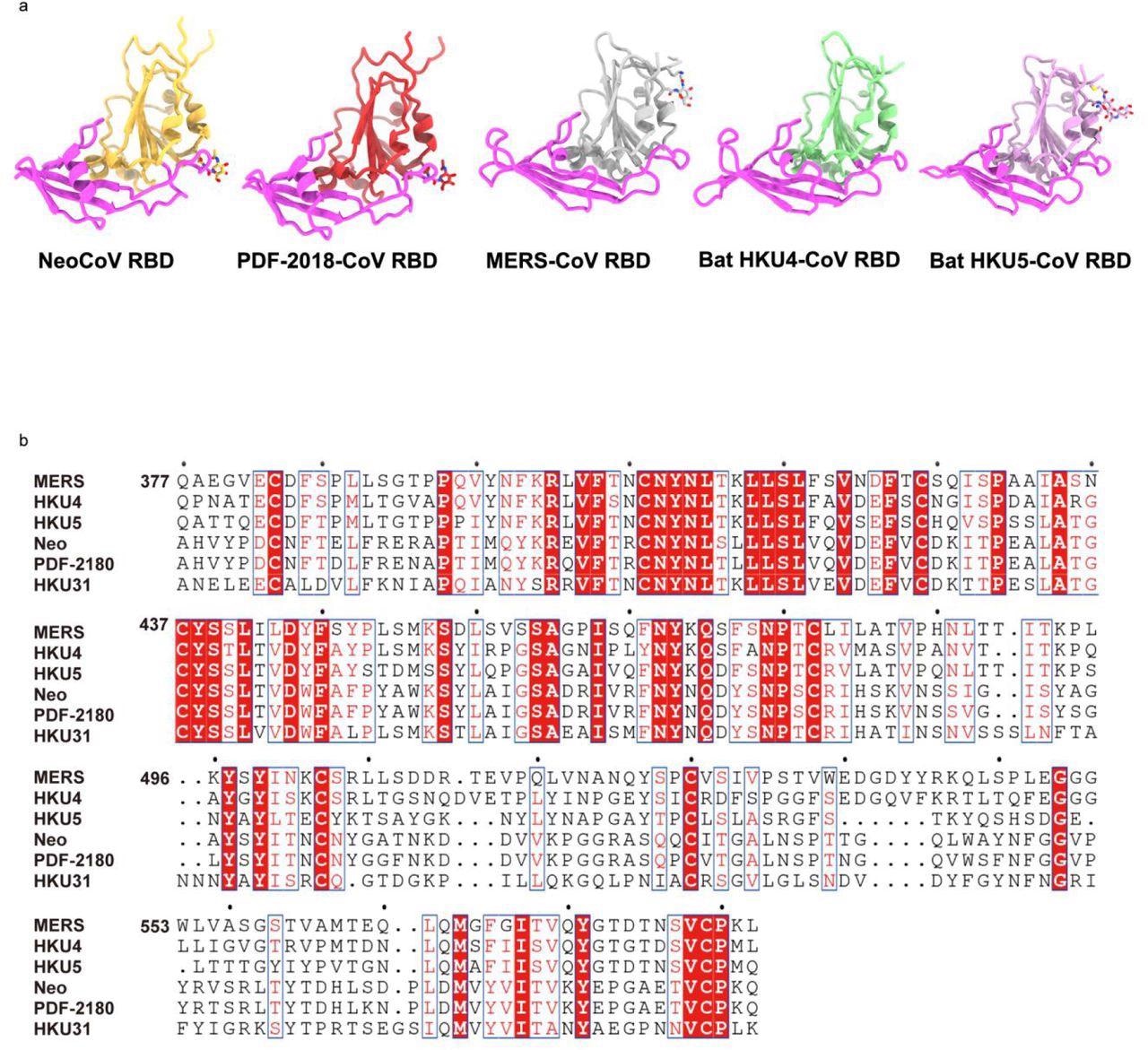
Structures and sequence comparison of RBDs from different merbecoviruses.
Conclusions
According to the authors, this is the first study demonstrating the ACE2 as a functional receptor in MERS-CoV-related viruses in bats. The ribonucleic acid (RNA) recombination during co-infection with different coronaviruses results in new viruses with distinct receptor usage and host tropisms.
The present findings support the previous hypothesis that MERS-CoV originated from the intra-S recombination between a DPP4-using virus and a NeoCoV-like virus. However, future studies are required to obtain sufficient evidence regarding the origin of MERS-CoV.
NeoCoV and PDF-2180-CoV did not show a significant affinity towards hACE2. However, a single residue substitution can increase local hydrophobicity near site 510 and enhance their hACE2-affinity, thus possessing a zoonotic potential in humans.
Overall, the current study emphasizes a potential biosafety threat of the emergence of MERS-CoV utilizing hACE2 associated with high mortality and transmission rates. The significance of this threat is further higher in light of the extensive mutations in the SARS-CoV-2 RBD regions, especially the heavily mutated Omicron variant.
Thus, appropriate surveillance and research on these two viruses are required to prepare healthcare systems for possible outbreaks of MERS-CoV with higher hACE2 affinity through antigenic drift in the future.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Xiong, Q., Cao, L., Ma, C., et al. (2022). Close relatives of MERS-CoV in bats use ACE2 as their functional receptors. bioRxiv. doi:10.1101/2022.01.24.477490. https://www.biorxiv.org/content/10.1101/2022.01.24.477490v1.
- Peer reviewed and published scientific report.
Xiong, Qing, Lei Cao, Chengbao Ma, M. Alejandra Tortorici, Chen Liu, Junyu Si, Peng Liu, et al. 2022. “Close Relatives of MERS-CoV in Bats Use ACE2 as Their Functional Receptors.” Nature 612 (7941): 748–57. https://doi.org/10.1038/s41586-022-05513-3. https://www.nature.com/articles/s41586-022-05513-3.