As the severe acute respiratory syndrome coronavirus 2 (SARS‑CoV‑2) Omicron variant makes its way across the world, it is one of five variants of concern that has made it challenging to end the pandemic.
While all variants evolved to have mutations that make them more virulent or deadly, they emerged independently of each other. Monitoring the genome of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is therefore essential for tracking any potential new variants.
A team of researchers led by Laura D. Hughes from The Scripps Research Institute in California created an online platform called Outbreak.info to track over 40 million lineages and SARS-CoV-2 mutation across 7,000 locations. This platform can help provide real-time genomic surveillance to help public officials respond to future public health threats.
The research paper Outbreak.info genomic reports: scalable and dynamic surveillance of SARS-CoV-2 variants and mutations” is recently published on the medRxiv* preprint server.
Study: Outbreak.info genomic reports: scalable and dynamic surveillance of SARS-CoV-2 variants and mutations. Image Credit: FOTOGRIN/Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Creating the platform
The researchers calculated the growth rate of a SARS-CoV-2 lineage based on how it spread and the mutations that allow a lineage to be more infectious. For instance, the Alpha variant was 40% to 60% more transmissible than prior lineages, and this biological feature allowed it to spread and become a dominant variant rapidly.
To maximize the full use of genomic surveillance data, the researchers created a platform called outbreak.info to search genomic data across three dimensions: geography, time, and lineages or mutations.
The team intentionally did not create a global phylogeny, instead relying on characteristic mutations to define a lineage. Doing so allowed for easier updating of a growing number of SARS-CoV-2 genomes.
The platform also collected epidemiological data from John Hopkins University and public literature and datasets from servers such as bioRxiv, medRxiv, and LitCOVID to assist genomic surveillance.
Genomic surveillance method using outbreak.info
Genomic data comes from the GISAID Initiative and is stored on a webserver. The web interface used three tools to parse genomic data: lineage and/or mutation tracker, location tracker, and lineage comparison tool. Each tool comes with an opinionated interface that focuses on one dimension of genomic data with the option to customize your search with more secondary dimensions.
The Lineage and/or Mutation tracker monitors the growth of a specific lineage, mutation, or a combination of the two. The tracker helps look at geographic variations of a variant of concern such as Delta and its sublineages as well as how it grew over time.
The Location Tracker focuses on a site for the potential SARS-CoV-2 lineage and the rate of mutations across various lineages in that given area. The researchers note this would be valuable information for studying changes in variants of concerns as most are regionally dominant. For instance, the Gamma variant is contained mainly in Brazil.
Tracking the growth of lineages at different geographic scales — at country, state/province, or county/city level — can help look at other factors influencing growth rates such as population immunity from vaccine or infection, social mobility, and viral attack rate. The Location Tracker surveys circulating lineages in a given area over the last 60 days and compares it to other circulating lineages in that location at different timepoints. Additionally, the tracker has information on how specific lineages affected caseloads in that area.
The Lineage Comparison Tool looks at the type of mutations across different lineages.
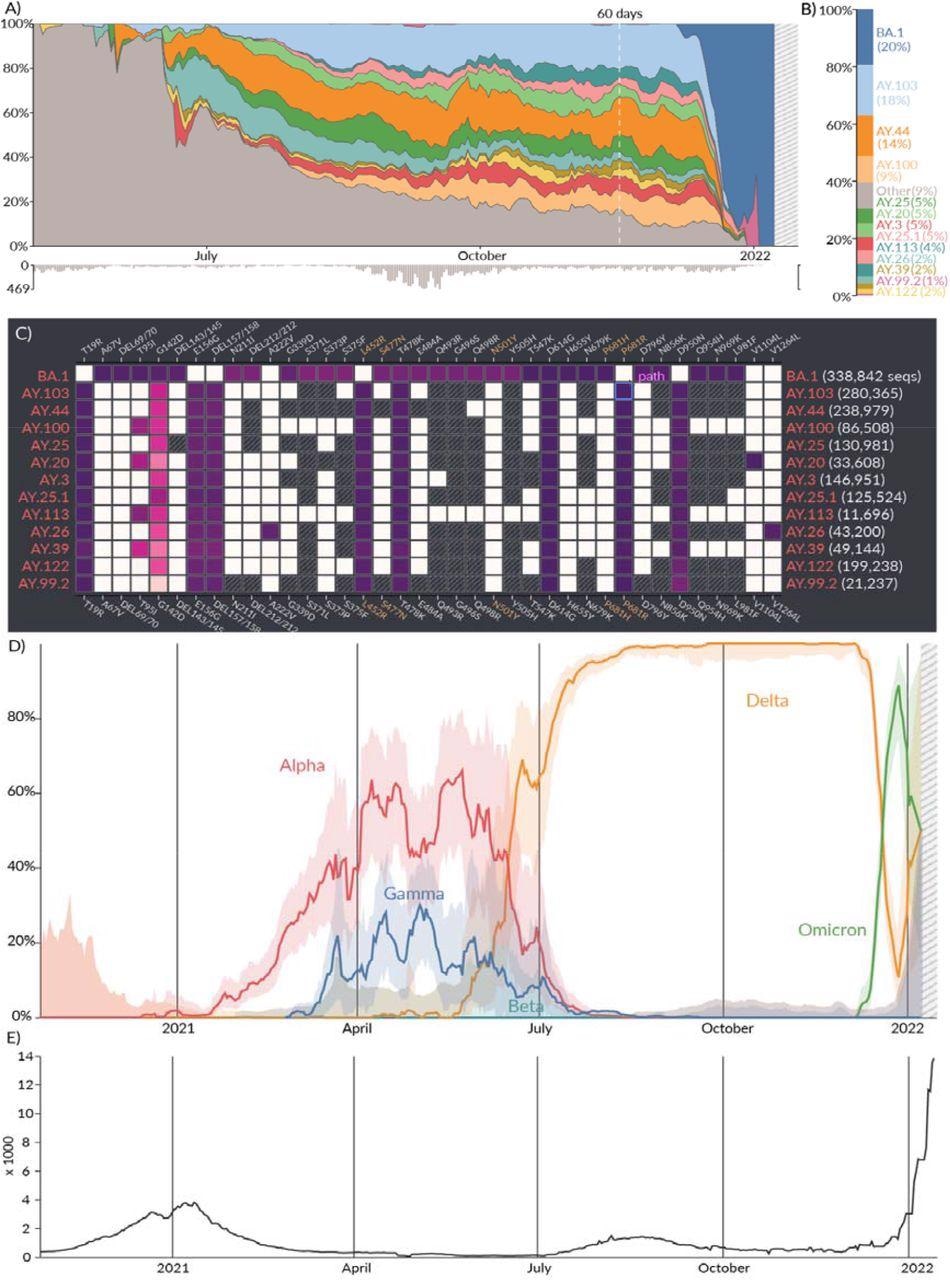
Location report. A) Relative prevalence of all lineages over time in San Diego, California. Total number of sequenced samples collected per day are shown in the bar chart below. B) Relative cumulative prevalence of all lineages over the last 60 days in San Diego. C) Mutation prevalence across the most prevalent lineages in San Diego over the last 60 days. D) Comparison of the prevalence of VOCs grouped by WHO classification: Alpha, Beta, Gamma, and Delta over time in San Diego. E) Daily reported cases in San Diego are shown in the line chart below.
Applying the platform to track and compare prevalence of VOC lineages
The Omicron lineage was first discovered in November 2021, where it overtook Delta as the dominant COVID-19 variant. Delta and Omicron both showed high growth rates with little variation worldwide.
A small percentage of the Alpha variant was circulating in Brazil and South Africa. However, Gamma and Beta were the dominant variants of concern at that time.
The Alpha variant was first detected in Southern England in late September 2020. Three months later, Alpha lineages appeared with a 40% to 50% increased transmission compared to prior lineages. The Alpha variant was dominant until the emergence of Delta in April 2021.
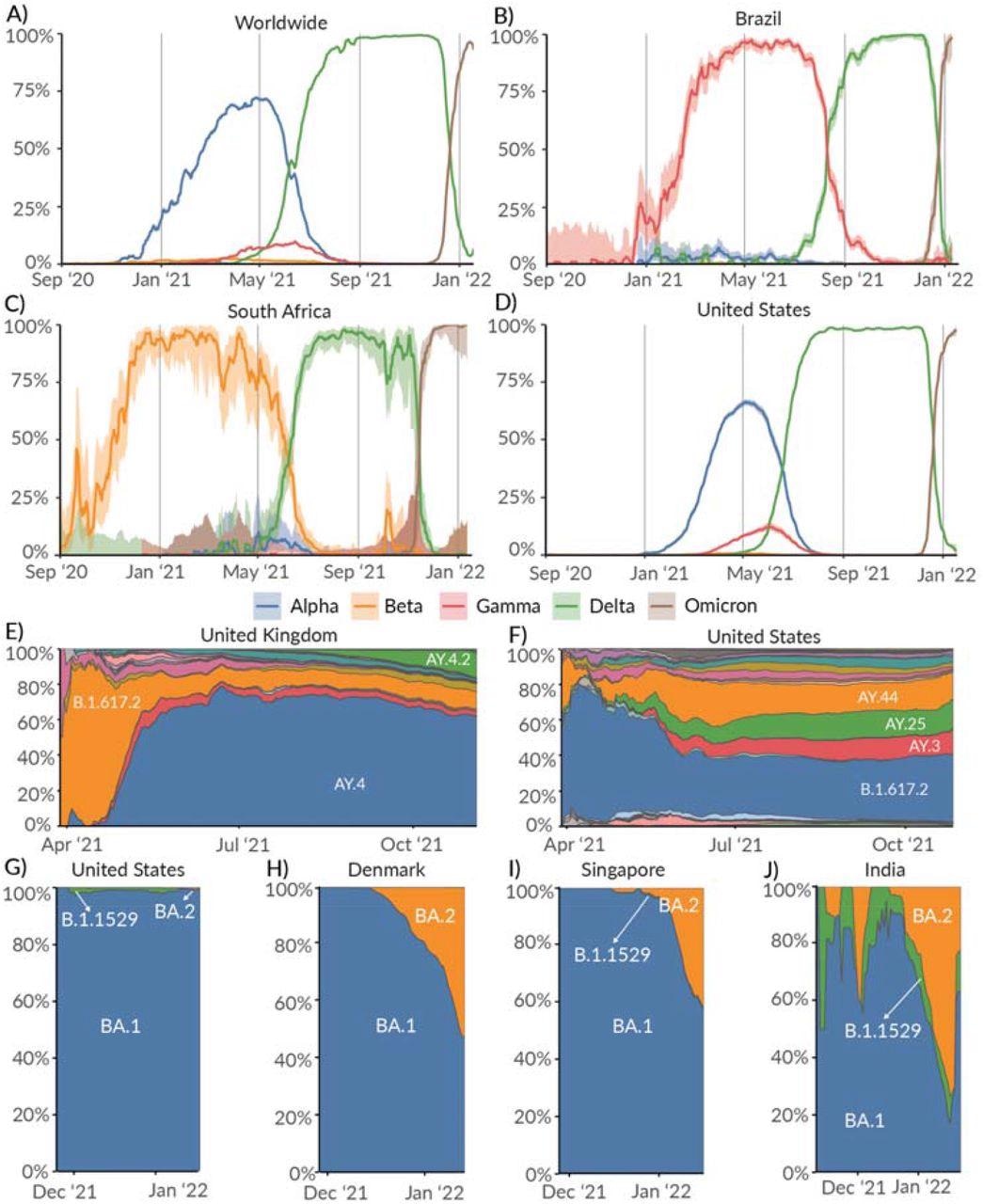
Prevalence of Variants of Concern: Alpha, Beta, Gamma, and Delta lineages over time in the A) Worldwide, B) Brazil, C) South Africa, and D) United States. Relative prevalence of sublineages within Delta variant over time in E) United Kingdom, and F) United States. Relative prevalence of sublineages within the Omicron variant over time in G) United States, H) Denmark, I) Singapore, and J) India.
Gamma made up 8% of U.S. COVID-19 cases. Further, coronavirus cases stemming from the Beta variant were less than 1%.
As Delta began to spread, it genetically diversified into more than 200 sublineages. The circulation of these sublineages depends on location with AY.4 most prevalent in the United Kingdom while B.1.617.2 was prevalent in the U.S.
The emergence of Omicron exhibited increased transmissibility than Delta because of mutations that enhance immune evasion, contributing to its rapid growth rate globally. BA.1 is the dominant sublineage of Omicron, but others belonging to BA.2 have been detected in Denmark, Singapore, and India.
“While there are no known phenotypic differences between BA.1 and BA.2, it remains to be seen if BA.2 will continue to grow in relative prevalence within Omicron globally,” wrote the researchers.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Gangavarapu K, et al. (2022). Outbreak.info genomic reports: scalable and dynamic surveillance of SARS-CoV-2 variants and mutations. medRxiv. Doi: https://doi.org/10.1101/2022.01.27.22269965, https://www.medrxiv.org/content/10.1101/2022.01.27.22269965v1
- Peer reviewed and published scientific report.
Gangavarapu, Karthik, Alaa Abdel Latif, Julia L. Mullen, Manar Alkuzweny, Emory Hufbauer, Ginger Tsueng, Emily Haag, et al. 2023. “Outbreak.info Genomic Reports: Scalable and Dynamic Surveillance of SARS-CoV-2 Variants and Mutations.” Nature Methods 20 (4): 512–22. https://doi.org/10.1038/s41592-023-01769-3. https://www.nature.com/articles/s41592-023-01769-3.