
 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
As new SARS-CoV-2 variants continually emerge, understanding the antigenic relationships among the variants from raw neutralization data is becoming increasingly complex. Despite these challenges, it remains crucial to understand these relationships among different SARS-CoV-2 variants to evaluate the need for vaccination updates and predict whether new variants will evade antibodies induced by current coronavirus disease 2019 (COVID-19) vaccines.
Background
Antigenic cartography is a core component of the bi-annual influenza virus vaccine strain selection process convened by the World Health Organization (WHO). Through the use of antigenic cartography, the researchers quantified and visualized the antigenic relationships among SARS-CoV-2 variants as antibody landscapes to demonstrate how reactivity clusters form in different regions of antigenic space.
About the study
The serum samples used in the current study were obtained from individuals who contracted SARS-CoV-2 infection with one of the seven different variants of SARS-CoV-2 or previously naïve individuals who had been vaccinated with the mRNA-1273 vaccine. Taken together, a total of 126 serum samples were used in a neutralization assay against a series of lentivirus pseudotypes.
For each sera group, geometric mean titers (GMT) and individual serum reactivity profiles were measured. In addition, the titration data of each sera group was used to construct an antigenic map in which the distance between the antigens and sera corresponded to the fold-drop compared to the maximum serum titer.
Study findings
As compared to D614G convalescent sera, post-vaccination sera exhibited fewer variations in regard to the magnitude of response and reactivity patterns against the SARS-CoV-2 variants. This is likely due to the more standardized dose and nature of vaccination as compared to post-infection sera.
Additional variations observed in post-infection serum groups were related to higher-than-expected titers against Delta (B.1.617.2)-like variants. This suggests that, in some cases, the patients with Delta-confirmed infections may have also experienced a previous infection with a different variant.
GMTs were consistently higher in post-vaccination sera. Furthermore, as compared to those who were infected with the homologous D614G strain, the fold-drop in GMT values was smaller for several variants in the vaccinated group. Taken together, these observations suggest that four weeks post-second dose, vaccine titers were not only higher than D614G convalescent titers, but the sera were also more cross-reactive.
Omicron showed the largest fold-decline in GMTs. This included an approximately 32-fold reduction against the post-vaccination and Beta sera, as well as an approximately 256-fold drop when tested against the Delta sera.
The antigenic map had six different areas that represented the clustering of different serum groups antigenically. In the antigenic map, the area occupied by the Omicron variant was positioned farthest from the vaccine and D614G convalescent sera.
The antigenic map showed individual serum responses as separate points based on fold-drop as compared to the maximum serum titer. This is in addition to depicting individual responses and mean estimates of the GMT for each serum group modeled as antibody landscape surfaces in a third dimension. Consistent with the findings based on raw neutralization titers, the antibody landscape illustrated that all tested serum groups had markedly reduced reactivity against B.1.1.529 (Omicron).
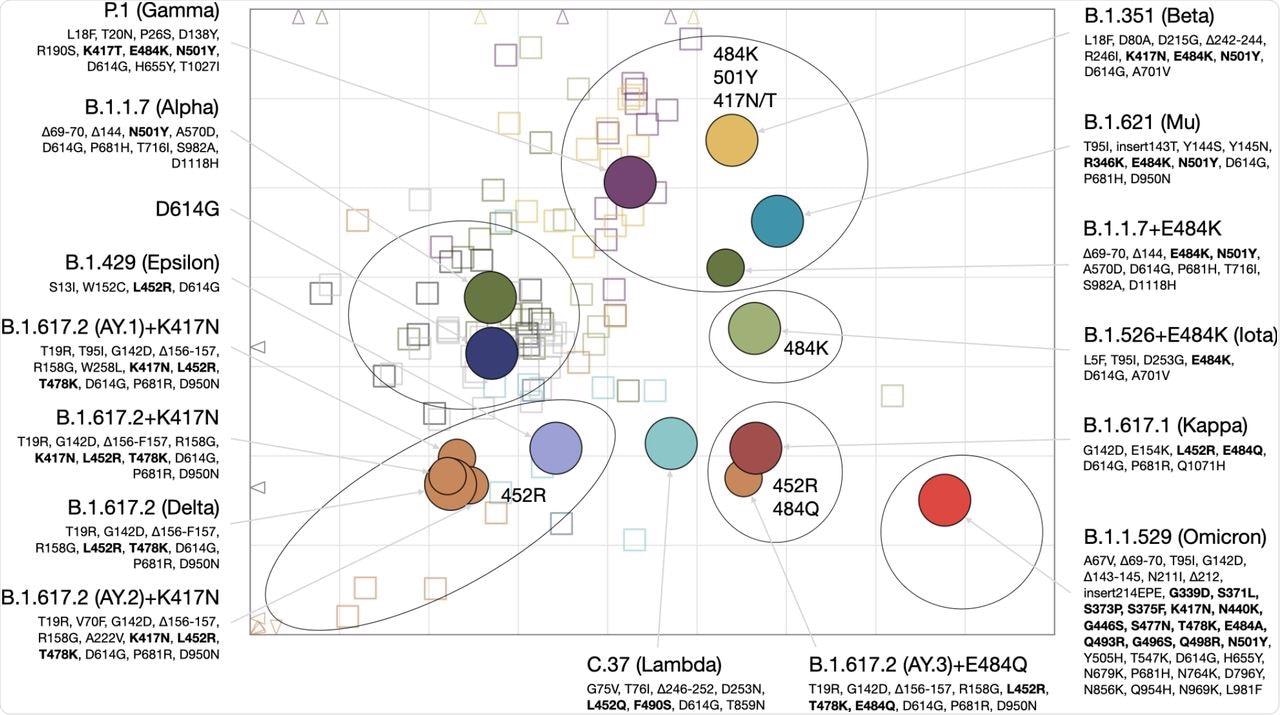
Antigenic map of SARS-CoV-2 variants and selected substitutions.
The researchers also observed switches in immunodominance among sera groups. The sera preferentially targeted different regions of the SARS-CoV-2 receptor-binding domains (RBD), depending on the variant to which an individual first got exposed.
These findings have potential consequences for future variant risk assessment and vaccine immunogen selection. These observations may also explain why the Alpha (B.1.1.7) variant with N501Y mutation in the RBD does not escape reactivity to a vaccine or D614G sera but can still be efficiently neutralized by B.1.351 sera.
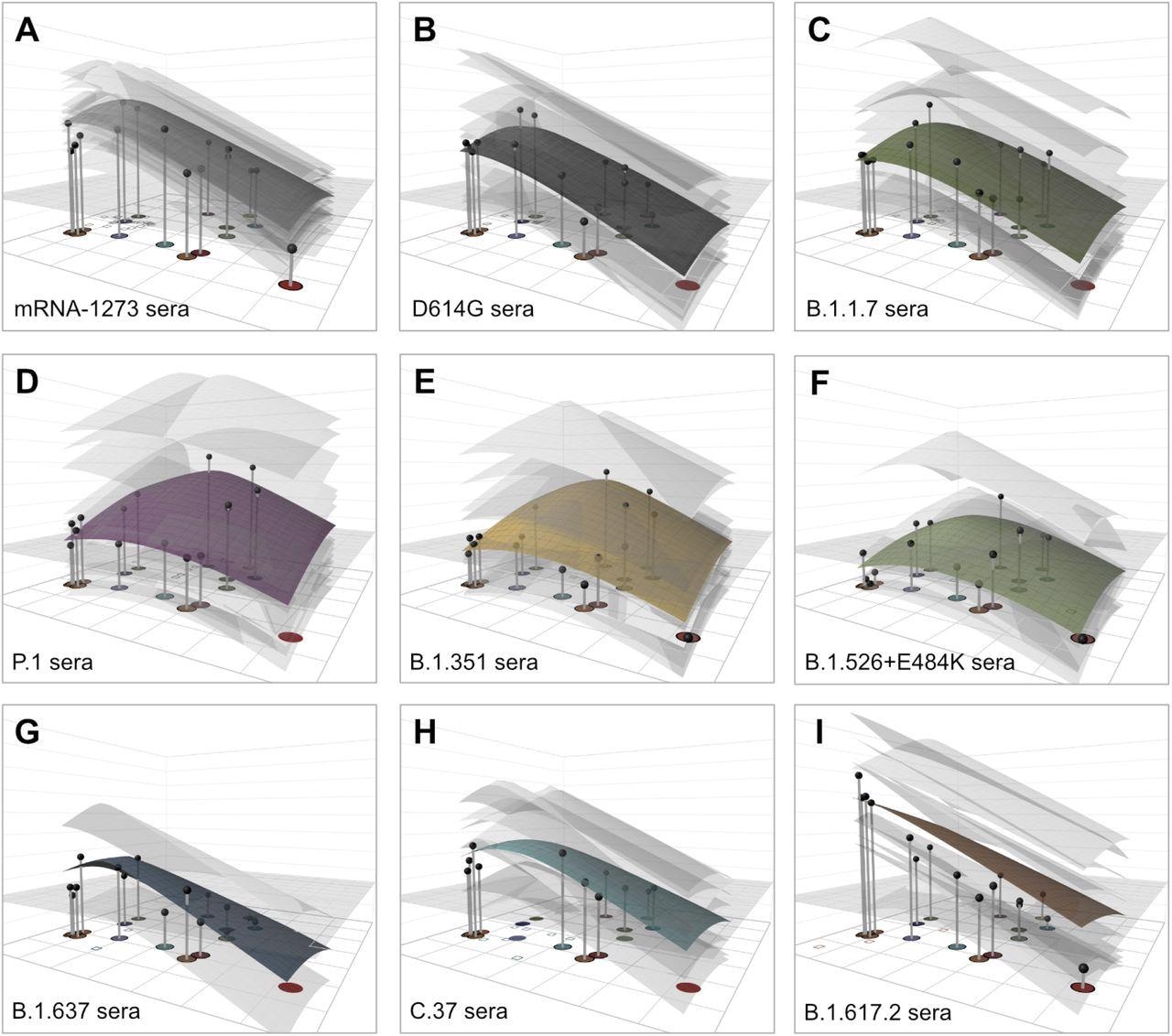
Antibody landscapes for each serum group. Colored surfaces show the GMT antibody landscapes for the different serum groups, light gray surfaces show the landscapes for each individual serum. Gray impulses show the height of the GMT for a specific variant, after accounting for individual effects as described in Materials and Methods (which would otherwise bias the GMT for variants not titrated against all sera). The base x-y plane corresponds to the antigenic map shown in Fig. 2. The vertical z-axis in each plot corresponds to the titer on the log2 scale, each two-fold increment is marked, starting from a titer of 20, one unit above the map surface. The gray horizontal plane indicates the height of a titer of 50, as a reference for judging the landscapes against various estimates of neutralizing antibody correlates of protection. Additional visualizations of predicted vs fitted titers are shown in fig. S21.
Conclusions
As populations will experience multiple exposures to different SARS-CoV-2 variants, studying the responses that consider these immunodominance patterns will be beneficial. Furthermore, responses to vaccination with new variants may eventually differ from the types of response inferred from first exposure sera and animal models, which may have targeted different antigenic epitopes.
With this knowledge, it will be possible for researchers to determine the variants and mutations to which different populations will be most vulnerable and help anticipate which emerging variants may evade current immunity.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.