Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of Coronavirus disease 2019 (COVID-19), wreaked havoc by mutating and circulating throughout the world.
Recently, COVID-19 has been profoundly affected by the emergence of the novel SARS-CoV-2 Omicron variant, B.1.1.529. The Omicron variant appears to be displacing Delta, becoming the dominant circulating variant, in most countries across the world.
Study: Spike Protein-independent Attenuation of SARS-CoV-2 Omicron Variant in Laboratory Mice. Image Credit: ktsdesign / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Studies have reported that the omicron variant is known to escape vaccine- or natural infection-elicited immunity, due to 30 plus mutations in its spike protein. The Omicron variant contains multiple amino acid changes within its receptor-binding domain (RBD), including S417N, T478K, E484A, Q493R, G496S, Q498R, N501Y, and Y505H, which may modify the virus's species tropism.
N501Y has been found in the Alpha (B1.1.7), Beta (B1.351), and P.1 variants and has been shown to improve spike binding to both human and mouse angiotensin-converting enzyme 2 (ACE2). The occurrence of N501Y, along with other mutations (K417M, E484K, Q493R, and Q498R) is suggested to be linked to serial passage adaption in mice. Notably, few recent studies reported that the Omicron variant often induces milder illnesses in experimental animals, often accompanied by a lower viral load.
In order to address this concern, a new study posted to the bioRxiv* preprint server provided the structural basis for a robust interaction between the receptor-binding domain of the Omicron spike protein and mouse ACE2. The study also compared the viral load and disease severity in laboratory mice infected with the natural Omicron variant, recombinant ancestral viruses bearing the Omicron spike protein, or, only the N501Y/Q493R mutations in the spike.
Findings
The lentiviral-based pseudoviral particles packaged with the bald virus (no spike protein), the ancestral WA1 spike, and the Omicron spike protein were allowed to transfect 293T cells with DNA plasmids to express the ACE2 of 10 species (human, pigs, ferrets, Syrian hamsters, mice, pangolins, horseshoe bats, bovines, African green monkeys, and Chinese hamsters).
It was found that most of the ACE2 homologs were equally capable of mediating the WA1- or Omicron-pseudovirus infection. However, mouse ACE2 (mACE2) expression enabled infections by the pseudovirus bearing the Omicron spike, but not by the WA1 spike. On the other hand, horseshoe bat ACE2 (hACE2), however, only mediated entry of the pseudovirus bearing the WA1 spike. Similar results were achieved when pseudoviruses encoding the firefly luciferase (FLuc) reporter gene were utilized in transient transfections or in stable hACE2 and mACE2 cell lines.
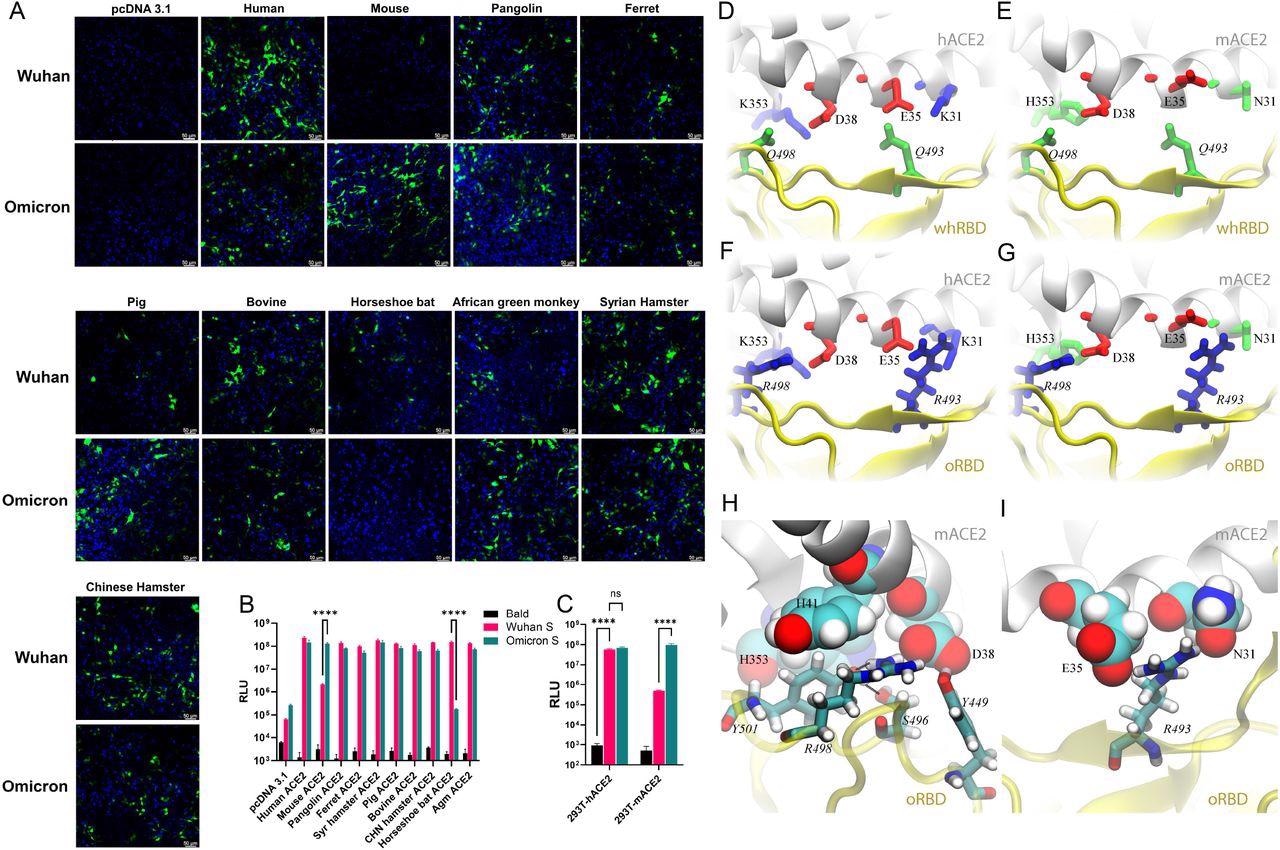
Pseudovirus bearing the Omicron spike protein efficiently infects cells expressing mACE2. (A) 293T cells were transfected with ACE2 expression plasmids of 10 species and then infected by lentiviral pseudoparticles bearing the spike protein of either the Wuhan SARS-CoV-2 variant or the Omicron variant. Infected cells would be GFP positive (green). Blue dye-stained nuclei. (B) A similar experiment was performed as in A except pseudoviruses expressing firefly luciferase were employed. (C) Infection of hACE2- and mACE2 stable 293T cells by pseudoviruses bearing the spike protein of either the Wuhan SARS-CoV-2 variant or the Omicron variant. (D—G) Various coordinations of charged residues at the ACE2-RBD interface. (D) hACE2-wtRBD (buried charge: 0), (E) mACE2-wtRBD (buried charge: −2 e), (F) hACE2-oRBD (buried charge: +2 e), (G) mACE2-oRBD (buried charge: 0). (H and I) Detailed interfacial coordinations between mACE2 and oRBD. (H) The interfacial binding near the salt-bridge R498-D38. (I)The interfacial binding near the salt-bridge R493-E35.
Further, the study employed molecular dynamics (MD) simulations in a physiology-like environment to recognize the structural basis of the mACE2-Omicron RBD (oRBD) interaction.
In comparison to the wild-type SARS-CoV-2 strain, the Omicron variant has 15 RBD mutations including the two charged alterations Q493R and Q498R, positioned inside the receptor-binding motif (RBM) that directly binds to ACE2. Two positively charged residues (R493 and R498) were found in the RBM of oRBD while two neutral residues Q493 and Q498 were present on the Wuhan-like ancestral RBD (whRBD) surface.
It was found that the two salt-bridges (K353-D38 and E35-K31) residing at the binding surface of hACE2 and two neutral residues, Q493 and Q498, present on the Wuhan-like ancestral RBD (whRBD) surface, formed interfacial hydrogen bonds. The two salt-bridges were partially buried after the hACE2-whRBD binding, which was energetically favorable.
When hACE2 was bound with oRBD , the net charge at the interface was +2 because both R493 and R498 were buried (partially) at the interface. This interfacial arrangement was energetically unfavorable and might weaken the interfacial binding, but the highly favorable N501Y mutation strengthened oRBD- hACE2 interaction.
Whereas, when negatively charged mACE2 was bound to whRBD, two negatively charged residues (D38 and E35), on the binding surface of mACE2, got buried at the binding interface, which was energetically unfavorable. When negatively charged, mACE2 electrostatically attracted positively charged oRBD, and the complex features of two salt bridges (R493-E35 and R498-D38) buried at the interface with zero net charge, which was energetically favorable, led to enhanced binding of mACE2 to oRBD.
The infection of 18-month-old Balb/c mice with the Omicron variant (isolate hCoV 19/USA/MD-HP20874/2021) was subsequently characterized. The study included a recombinant ancestral virus (WA1) bearing the entire Omicron Spike (WA1-Omicron-S) and a positive control Q493R/N501Y (WA1-Q493R/N501Y).
WA1-Omicron-S and Q493R/N501Y-infected mice developed clinical symptoms three- and four days post-infection (DPI), and the viral loads identified in infected mice were two logs higher when compared to naturally infected animals. The infection also significantly increased the interferon-stimulated gene 15 (ISG15) expression. Histopathology of the natural Omicron variant infected lung tissues revealed that the majority of infected animals had minimal immune cell infiltration. Whereas, infected mice with WA1-Omicron-S and WA1-Q493R/N501Y had extensive alveolar inflammatory infiltrates.
One possible limitation of the current work is limited inoculum, including the fact that only one natural Omicron variation was investigated. Because of the minimal inoculum, the study groups may have experienced mild disease overall.
Implications
Conclusively, these results suggest that, despite the Omicron variant’s efficient usage of mACE2, genetic changes outside the spike protein in the natural Omicron version contribute to the observed attenuation in laboratory mice.
Further, the low viral load reported in Omicron-infected mice is unlikely due to the variant’s inability to enter mouse tissues, however, alterations within the spike protein may still influence disease severity. The current study demonstrated that decreased viral load and attenuation observed in laboratory animals relate to post-entry blocking caused by mutations outside the spike protein in the Omicron variant.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources